Published online Apr 28, 2006. doi: 10.3748/wjg.v12.i16.2536
Revised: December 1, 2005
Accepted: December 7, 2005
Published online: April 28, 2006
AIM: To assess the effects of obstructive cholestasis on a wider range of gene expression using microarray technology.
METHODS: Male C57BL/6J mice underwent common bile duct ligation (BDL) and were matched with pair-fed sham-operated controls. After 7 d, the animals were sacrificed and total RNA was isolated from livers and kidneys. Equal amounts of RNA from each tissue were pooled for each group and hybridized to Affymetrix GeneChip®MG-U74Av2 containing a total of 12 488 probe sets. Data analysis was performed using GeneSpring®6.0 software. Northern analysis and immunofluorescence were used for validation.
RESULTS: In sham-operated and BDL mice, 44 and 50% of 12 488 genes were expressed in livers, whereas 49 and 51% were expressed in kidneys, respectively. Seven days after BDL, 265 liver and 112 kidney genes with GeneOntology annotation were up-regulated and 113 liver and 36 kidney genes were down-regulated in comparison with sham-operated controls. Many genes were commonly regulated in both tissues and metabolism-related genes represented the largest functional group.
CONCLUSION: Following BDL, microarray analysis reveals a broad range of gene alterations in both liver and kidney.
- Citation: Denk GU, Cai SY, Chen WS, Lin A, Soroka CJ, Boyer JL. A comparison of gene expression in mouse liver and kidney in obstructive cholestasis utilizing high-density oligonucleotide microarray technology. World J Gastroenterol 2006; 12(16): 2536-2548
- URL: https://www.wjgnet.com/1007-9327/full/v12/i16/2536.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i16.2536
Cholestasis, defined as impairment of bile secretion, is a feature of many hepatic disorders and systemic diseases. The recent cloning and functional characterization of different transport proteins for bile acids, organic anions and cations in hepatocytes and cholangiocytes have provided new insights into the molecular biology and physiology of bile formation and have increased understanding of the pathophysiology of cholestatic disorders[1]. Thus it is now established that a number of transport proteins in the basolateral and canalicular hepatocyte membrane undergo adaptive regulation in response to cholestatic liver injury to minimize the hepatic accumulation of toxic substances, such as hydrophobic bile acids[2-4]. Previous studies have indicated that in addition to the liver, adaptive regulation of these transporters in cholestasis also occurs in extrahepatic tissues, including the kidney[5] and the intestine[6]. Other alterations in cholestasis affect hepatic signal transduction[7,8], vesicular transport[7], apoptosis[9,10], metabolism[11], and the structure of the extracellular matrix[12,13].
Given the wide range of signaling, regulatory, and metabolic pathways, structural elements, and transport proteins which may be affected in cholestasis, much further research will be necessary to more fully understand the extent of these adaptations. High-density DNA microarrays containing thousands of DNA fragments and oligonucleotides are a potentially promising approach to identify additional genes of interest that play a role in this pathophysiologic process. Based on their ability to monitor large numbers of genes at a time, high-density DNA microarrays are a sensitive, time-saving, and efficient tool in determining gene expression and finding regulatory pathways[14].
In the present study, we, therefore, have utilized high-density oligonucleotide microarray technology to screen for gene alterations in the liver and kidney following bile duct ligation (BDL) in mice, an established model of obstructive cholestasis. This study has allowed a comprehensive gene expression profile to be obtained in cholestatic mouse liver and kidney as well as it has highlighted a number of genes whose expression is particularly altered by this process.
Male C57BL/6J mice (8-12-wk-old) purchased from Jackson Lab (Bar Harbor, ME) underwent BDL or sham-surgery as previously described[15]. The common bile duct was identified, ligated twice close to the liver hilum immediately below the cystic duct, and then divided between the ligatures. Control mice underwent sham-surgery in which the common bile duct was exposed but not ligated. Since sham-operated mice tend to consume more food than BDL mice and the expression of some genes may be affected by caloric intake, food intake of BDL mice was monitored daily and sham-operated mice were pair-fed so as to receive the same amount of food as BDL mice. Animals were sacrificed 7 d after surgery and livers and kidneys were harvested. The protocol was approved by the Yale Animal Care and Use Committee, and the animals received humane care as outlined in the “Guide for the Care and Use of Laboratory Animals” (NIH publication 86-23, revised 1985).
Blood-free livers and kidneys were homogenized in GTC solution containing 4 mol/L guanidinium thiocyanate, 25 mmol/L Na-citrate, and 5 g/L N-lauroylsarcosine and subjected to CsCl gradient centrifugation. The recovered total RNA was further purified by phenol/chloroform extraction and ethanol precipitation. The RNA concentration was determined spectrophotometrically and the RNA quality was confirmed by formaldehyde-agarose gel electrophoresis. Equal amounts of liver and kidney, respectively, total RNA from each of four BDL and four sham-operated mice were pooled to minimize inter-animal variations and used for biotin-labeling.
The biotin-labeled RNA from the different groups was hybridized with two replicates for each condition to individual high-density oligonucleotide microarray chips (GeneChip®MG-U74Av2) from Affymetrix (Santa Clara, CA) containing a total of 12 488 probe sets. Microarray expression data were generated with Affymetrix Microarray Suite 5.0 software and further analysis was carried out with GeneSpring®6.0 (Silicon Genetics, Redwood City, CA). Raw intensity values from each chip were normalized to the 50th percentile of the measurements taken from that chip to reduce chip-wide variations in intensity. Each gene was normalized to the average measurement of that gene in the respective paired controls to enable comparison of relative changes in gene expression levels between different conditions. Cross-gene error model was active based on the replicates. Comparisons of gene expression data were made between BDL and sham-operated mice. Signal and detection flag from Microarray Suite 5.0 were used as quality controls. Only genes with a minimum signal intensity of 600, a detection flag present in both replicates in at least one of the comparison conditions, and a two-fold and above change in gene expression were used for further analysis. For identification of differentially expressed genes in the different groups, a one-sample t-test with a P value cutoff of 0.05 was performed to determine if the average log of the ratio of the replicates was significantly different from 1.0, which was the value of the control samples after normalization. Finally, genes were categorized into GeneOntology (GO) and annotated using NetAffx®, an analysis web interface from Affymetrix.
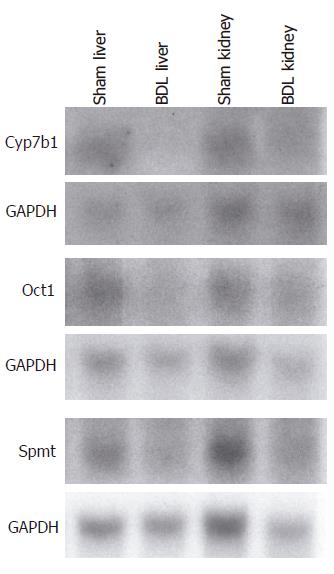
To validate alterations in gene expression on the microarray, changes in the expression of selected genes were confirmed in aliquots of the same RNA samples used for the microarray by Northern analysis as previously described[16]. The following primers were used for the generation of specific probes: cytochrome P450 7b1 (GenBank accession number U36993): 5’-GAATCTCAGCTTAGAGAGTAAGAG-3’ (sense), 5’-TTTGTACCTAAAGGAGACGGCAG-3’ (antisense); organic cation transporter 1 (Oct1) (GenBank accession number U38652): 5’-GCAGCCTGCCTCCTCATGATC-3’ (sense), 5’-GGTAAATCGTGTTTTCTTTGGCC-3’ (antisense); similar to putative integral membrane transport protein (GenBank accession number AI647632): 5’-TGATTACAAGAAATGTCAAGCAGG-3’ (sense), 5’-CCTCTTCCTGACTCCATCCATG-3’ (antisense).
Of the total of 12 488 genes on the microarray chip, 44 and 50% were expressed in the livers of sham-operated and BDL mice, respectively. After 7 d of obstructive cholestasis 265 genes with GO annotation were up-regulated and 113 were down-regulated in livers of BDL mice by a factor of two or more in comparison with sham-operated pair-fed controls. Metabolism-related genes represented the largest functional group among the altered genes after BDL in liver (Table 1). It should be noted that the grouping of the altered genes was primarily done to achieve a clearer arrangement for the reader. Since a considerable number of the encoded proteins have multiple, little characterized or even unknown functions, we want to point out that the classification provided is subject to the personal opinions and emphasis of the authors (Table 1). Upon request, a complete list of the altered genes including genes without GO annotation that are not mentioned here can be obtained from the authors. Alternatively, the complete list of altered genes can be accessed via http://livercenter.yale.edu/datalist.html.
| Liver | Kidney | Accession number | Description |
| Cell death | |||
| 3.6 | AF011428 | CD5 antigen-like | |
| 3.2 | AW046181 | Serum/glucocorticoid regulated kinase | |
| 2.6 | AV373612 | Bcl2-associated athanogene 3 | |
| -2.6 | X65128 | Growth arrest specific 1 | |
| -2.7 | AA770736 | Induced in fatty liver dystrophy 2 | |
| 2.8 | M61737 | Fat-specific gene 27 | |
| 2.7 | AV003873 | Clusterin | |
| 2.3 | D14077 | Clusterin | |
| -7 | AJ000062 | Deoxyribonuclease I | |
| Stress response | |||
| 10 | 36.1 | X03505 | Serum amyloid A 3 |
| 5 | M13521 | Serum amyloid A 2 | |
| 2.8 | M12566 | Orosomucoid 2 | |
| 2.5 | J04633 | Heat shock protein 1, alpha | |
| 2.5 | X60676 | Serine (or cysteine) proteinase inhibitor, clade H, member 1 | |
| 7.4 | M96827 | Haptoglobin | |
| -2.2 | Z36774 | Serine (or cysteine) proteinase inhibitor, clade F, member 2 | |
| Immune and inflammatory response | |||
| 31.9 | 5 | M94584 | Chitinase 3-like 3 |
| 17.3 | M19681 | Chemokine (C-C motif) ligand 2 | |
| 10.7 | X53798 | Chemokine (C-X-C motif) ligand 2 | |
| 9.6 | 4.2 | J04596 | Chemokine (C-X-C motif) ligand 1 |
| 9.4 | AW120786 | Chemokine (C-X-C motif) ligand 14 | |
| 9.2 | U18424 | Macrophage receptor with collagenous structure | |
| 8.4 | AV370035 | Chemokine (C-C motif) receptor 5 | |
| 7.1 | U56819 | Chemokine (C-C) receptor 2 | |
| 6.9 | 5.5 | AF002719 | Secretory leukocyte protease inhibitor |
| 6.1 | M18237 | Immunoglobulin kappa chain variable 8 (V8) | |
| 5.9 | U34277 | Phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | |
| 5.4 | M83218 | S100 calcium binding protein A8 (calgranulin A) | |
| 3.6 | 3.7 | X04673 | Adipsin |
| 3.6 | AI844520 | Interferon gamma inducible protein 30 | |
| 3.6 | AF081789 | Complement component 1, q subcomponent, receptor 1 | |
| 3.5 | X12905 | Properdin factor, complement | |
| 3.3 | L32838 | Interleukin 1 receptor antagonist | |
| 3.2 | U96752 | Histocompatibility 2, Q region locus 1 | |
| 3.2 | 3.4 | M22531 | Complement component 1, q subcomponent, beta polypeptide |
| 3.2 | X15591 | Cytotoxic T lymphocyte-associated protein 2 alpha | |
| 3.1 | X63782 | Lymphocyte antigen 6 complex, locus D | |
| 2.9 | M58004 | Chemokine (C-C motif) ligand 6 | |
| 2.9 | M21932 | Histocompatibility 2, class II antigen A, beta 1 | |
| 2.9 | U16985 | Lymphotoxin B | |
| 2.9 | M14639 | Interleukin 1 alpha | |
| 2.9 | 4.2 | X58861 | Complement component 1, q subcomponent, alpha polypeptide |
| 2.8 | U77461 | Complement component 3a receptor 1 | |
| 2.8 | M31314 | Fc receptor, IgG, high affinity I | |
| 2.8 | X52643 | Histocompatibility 2, class II antigen A, alpha | |
| 2.7 | AB007599 | Lymphocyte antigen 86 | |
| 2.6 | X15592 | Cytotoxic T lymphocyte-associated protein 2 beta | |
| 2.6 | AF013715 | Periplakin | |
| 2.6 | 2.1 | X66295 | Complement component 1, q subcomponent, gamma polypeptide |
| 2.5 | 3.1 | L38444 | T-cell specific GTPase |
| 2.5 | AA596710 | Leukotriene B4 12-hydroxydehydrogenase | |
| 2.5 | AB019505 | Interleukin 18 binding protein | |
| 2.4 | M34815 | Chemokine (C-X-C motif) ligand 9 | |
| 2.4 | 2 | X00496 | Ia-associated invariant chain |
| 2.4 | 2.8 | AJ007970 | Guanylate nucleotide binding protein 2 |
| 2.3 | D86382 | Allograft inflammatory factor 1 | |
| 2.3 | L22181 | Formyl peptide receptor 1 | |
| 2.2 | AF038149 | Paired-Ig-like receptor B | |
| 2.1 | AW060457 | Immunoglobulin superfamily, member 7 | |
| 2.1 | U03003 | Defensin related cryptdin 6 | |
| 2 | M29855 | Colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) | |
| 2 | AF003525 | Defensin beta 1 | |
| -2.8 | M29007 | Complement component factor h | |
| -4.6 | L22977 | X-linked lymphocyte-regulated 3b | |
| 13.6 | U47810 | Complement component factor i | |
| 6.5 | K02782 | Complement component 3 | |
| 6 | X06454 | Complement component 4 (within H-2S) | |
| 4.2 | AI563854 | Tumor-associated calcium signal transducer 2 | |
| 3.8 | AA986114 | T-cell immunoglobulin and mucin domain containing 2 | |
| 3.5 | U49513 | Chemokine (C-C motif) ligand 9 | |
| 3 | Y08830 | Tumor-associated calcium signal transducer 2 | |
| 2.4 | AA270365 | Cytokine receptor-like factor 1 | |
| 2.2 | AI152789 | Sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A | |
| Signal transduction | |||
| 12.9 | U88328 | Suppressor of cytokine signaling 3 | |
| 5.4 | Z48043 | Coagulation factor II (thrombin) receptor-like 1 | |
| 5 | 2.7 | M14044 | Annexin A2 |
| 4 | 2.2 | AJ001633 | Annexin A3 |
| 3.6 | AI641895 | Shroom | |
| 3.6 | U90715 | Coxsackievirus and adenovirus receptor | |
| 3.6 | AI317205 | Mitogen activated protein kinase kinase kinase 1 | |
| 3.4 | J03023 | Hemopoietic cell kinase | |
| 3.1 | AW209098 | IQ motif containing GTPase activating protein 1 | |
| 3 | AW049806 | RIKEN cDNA 1700093E07 gene | |
| 3 | X84797 | Hematopoietic cell specific Lyn substrate 1 | |
| 2.9 | 3 | AB015978 | Oncostatin M receptor |
| 2.6 | X93328 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 | |
| 2.3 | D63423 | Annexin A5 | |
| 2.3 | 2.1 | M69260 | Annexin A1 |
| 2.2 | M68902 | Hemopoietic cell phosphatase | |
| 2.2 | AF020313 | Amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | |
| 2.1 | 3.6 | AV374868 | Suppressor of cytokine signaling 3 |
| 2.1 | AA608387 | Interleukin 13 receptor, alpha 1 | |
| -2 | AC002397 | Gene rich cluster, C9 gene | |
| -2 | AW125649 | Guanine nucleotide binding protein, alpha 12 | |
| -2.4 | AI839138 | Thioredoxin interacting protein | |
| -2.6 | AV321519 | Sorting nexin 17 | |
| -2.7 | AA691492 | RIKEN cDNA D530020C15 gene | |
| -5.6 | D17444 | Leukemia inhibitory factor receptor | |
| -11.7 | AV349152 | Regulator of G-protein signaling 16 | |
| -15.3 | U94828 | Regulator of G-protein signaling 16 | |
| 2.3 | AF084466 | Ras-related associated with diabetes | |
| 2.1 | AF009246 | RAS, dexamethasone-induced 1 | |
| -2.1 | AF054623 | Frizzled homolog 1 (Drosophila) | |
| -2.2 | D85605 | Cholecystokinin A receptor | |
| -2.2 | AI834895 | Membrane progestin receptor alpha | |
| -2.3 | AW046638 | PDZ domain containing 1 | |
| Cell growth and maintenance | |||
| 8.7 | M33960 | Serine (or cysteine) proteinase inhibitor, clade E, member 1 | |
| 6.9 | X98471 | Epithelial membrane protein 1 | |
| 5.8 | 4.9 | X66449 | S100 calcium binding protein A6 (calcyclin) |
| 5.4 | AF055638 | Growth arrest and DNA-damage-inducible 45 gamma | |
| 5.1 | M17298 | Nerve growth factor, beta | |
| 3.6 | AI849928 | Cyclin D1 | |
| 3.5 | X59846 | Growth arrest specific 6 | |
| 3.2 | M64292 | B-cell translocation gene 2, anti-proliferative | |
| 3.2 | AW048937 | Cyclin-dependent kinase inhibitor 1A (P21) | |
| 3.1 | AF009366 | Neural precursor cell expressed, developmentally down-regulated gene 9 | |
| 2.7 | M21019 | Harvey rat sarcoma oncogene, subgroup R | |
| 2.7 | X06368 | Colony-stimulating factor 1 receptor | |
| 2.2 | X81579 | Insulin-like growth factor binding protein 1 | |
| 2.1 | AI851454 | Cysteine rich protein 2 | |
| 2 | AA529583 | Mortality factor 4 like 2 | |
| -2.1 | X95280 | G0/G1 switch gene 2 | |
| -2.2 | M31680 | Growth hormone receptor | |
| -2.5 | U15012 | Growth hormone receptor | |
| 3.4 | AI852641 | Nuclear protein 1 | |
| 2.8 | M34094 | Midkine | |
| 2.8 | AF058798 | Stratifin | |
| 2.1 | X81580 | Insulin-like growth factor binding protein 2 | |
| Protein biosynthesis | |||
| 2.3 | Y11460 | Integrin beta 4 binding protein | |
| 2.1 | NM_011690 | Valyl-tRNA synthetase 2 | |
| -2 | AV055186 | Ribosomal protein, large, P1 | |
| Proteolysis and protein degradation | |||
| 7.6 | 2.4 | X61232 | Carboxypeptidase E |
| 6.1 | AW060527 | Ubiquitin-conjugating enzyme E2 variant 2 | |
| 4 | AJ000990 | Legumain | |
| 4 | 5 | AJ223208 | Cathepsin S |
| 3.7 | AL078630 | Ubiquitin D | |
| 2 | U35833 | Ubiquitin-like 1 (sentrin) activating enzyme E1B | |
| -2.2 | AI844932 | F-box only protein 8 | |
| -2.4 | L21221 | Proprotein convertase subtilisin/kexin type 4 | |
| -2.6 | AV359471 | Ubiquitin specific protease 15 | |
| -2.2 | J04946 | Angiotensin converting enzyme | |
| -2.5 | L15193 | Meprin 1 beta | |
| Protein amino acid phosphorylation and dephosphorylation | |||
| 3.2 | D89728 | Serine/threonine kinase 10 | |
| 3.2 | M97590 | Protein tyrosine phosphatase, non-receptor type 1 | |
| 2.6 | D37801 | Protein tyrosine phosphatase, non-receptor type 21 | |
| 2 | X61940 | Dual specificity phosphatase 1 | |
| -2.1 | L31783 | Uridine monophosphate kinase | |
| Cell adhesion and extracellular matrix | |||
| 24.7 | L36244 | Matrix metalloproteinase 7 | |
| 20.6 | U43525 | Proteinase 3 | |
| 10.7 | M82831 | Matrix metalloproteinase 12 | |
| 10.4 | 2.3 | D00613 | Matrix gamma-carboxyglutamate (gla) protein |
| 9 | 3.1 | X16834 | Lectin, galactose binding, soluble 3 |
| 8.9 | L02918 | Procollagen, type V, alpha 2 | |
| 8.1 | M31039 | Integrin beta 2 | |
| 7.2 | M62470 | Thrombospondin 1 | |
| 6.2 | X13986 | Secreted phosphoprotein 1 | |
| 5.9 | 2.4 | U03419 | Procollagen, type I, alpha 1 |
| 5.8 | D14010 | Regenerating islet-derived 1 | |
| 4.9 | 2.1 | X52046 | Procollagen, type III, alpha 1 |
| 4.7 | 2.5 | M90551 | Intercellular adhesion molecule |
| 4.2 | X58251 | Procollagen, type I, alpha 2 | |
| 4 | L57509 | Discoidin domain receptor family, member 1 | |
| 3.4 | 4.3 | U12884 | Vascular cell adhesion molecule 1 |
| 3.2 | 3.3 | M84487 | Vascular cell adhesion molecule 1 |
| 3.2 | L29454 | Fibrillin 1 | |
| 3 | Z22532 | Syndecan 1 | |
| 2.9 | M23552 | Serum amyloid P-component | |
| 2.8 | X04017 | Secreted acidic cysteine rich glycoprotein | |
| 2.7 | M38337 | Milk fat globule-EGF factor 8 protein | |
| 2.7 | AA763466 | Procollagen, type I, alpha 1 | |
| 2.5 | AA919594 | Elastin | |
| 2.5 | M70642 | Connective tissue growth factor | |
| 2.5 | D88577 | C-type (calcium dependent, carbohydrate recognition domain) lectin, superfamily member 13 | |
| 2.3 | M15832 | Procollagen, type IV, alpha 1 | |
| 2.2 | X59990 | Catenin alpha 1 | |
| 2.2 | U82624 | Amyloid beta (A4) precursor protein | |
| 2.1 | X53928 | Biglycan | |
| 2.1 | U89915 | F11 receptor | |
| 2 | X04647 | Procollagen, type IV, alpha 2 | |
| 2 | 2.2 | V00755 | Tissue inhibitor of metalloproteinase 1 |
| 2 | X91144 | Selectin, platelet (p-selectin) ligand | |
| -2.1 | AF101164 | CEA-related cell adhesion molecule 2 | |
| -2.2 | AI840501 | Camello-like 1 | |
| 2.1 | L19932 | Transforming growth factor, beta induced | |
| Cytoskeleton and structural elements | |||
| 5 | 7.3 | M36120 | Keratin complex 1, acidic, gene 19 |
| 4.8 | V00830 | Keratin complex 1, acidic, gene 10 | |
| 4.5 | U38967 | Thymosin, beta 4, X chromosome | |
| 3.6 | 2.3 | AI852553 | Thymosin, beta 10 |
| 3.6 | U42471 | Wiskott-Aldrich syndrome homolog (human) | |
| 3.4 | U29539 | Lysosomal-associated protein transmembrane 5 | |
| 3.4 | M22479 | Tropomyosin 1, alpha | |
| 3.2 | 2.2 | M28739 | Tubulin, beta 2 |
| 3.2 | AW215736 | RIKEN cDNA 2310057H16 gene | |
| 3.2 | 4.5 | M22832 | Keratin complex 1, acidic, gene 18 |
| 3.1 | AI505453 | Myosin heavy chain IX | |
| 3 | X15662 | Keratin complex 2, basic, gene 8 | |
| 2.8 | X60671 | Villin 2 | |
| 2.7 | D49733 | Lamin A | |
| 2.7 | AW125446 | Golgi phosphoprotein 2 | |
| 2.6 | AW050256 | Tubulin, beta 3 | |
| 2.6 | AI839417 | Moesin | |
| 2.4 | AW125698 | Myosin heavy chain IX | |
| 2.4 | AW212775 | Actin-related protein 2/3 complex, subunit 1B | |
| 2.4 | AV356071 | Lysosomal-associated protein transmembrane 5 | |
| 2.2 | M28727 | Tubulin, alpha 2 | |
| 2.2 | AI835858 | Tropomyosin 4 | |
| 2.2 | M12347 | Actin, alpha 1, skeletal muscle | |
| 2.1 | D88793 | Cysteine and glycine-rich protein 1 | |
| 2.1 | AF020185 | Dynein, cytoplasmic, light chain 1 | |
| 2.1 | AI837625 | Cysteine and glycine-rich protein 1 | |
| 2.1 | 3.2 | X54511 | Capping protein (actin filament), gelsolin-like |
| 2 | 2.1 | X04663 | Tubulin, beta 5 |
| 2 | AI841606 | Actin-binding LIM protein 1 | |
| 2 | M21495 | Actin, gamma, cytoplasmic | |
| 2 | AI849152 | Clathrin, light polypeptide (Lcb) | |
| 2 | M60474 | Myristoylated alanine rich protein kinase C substrate | |
| -2.2 | AW123904 | Gamma-aminobutyric acid (GABA(A)) receptor-associated protein-like 1 | |
| 3.3 | AB000713 | Caudin 4 | |
| 2.6 | AA755126 | Keratin complex 2, basic, gene 7 | |
| 2.6 | AF087825 | Claudin 7 | |
| 2.3 | AI195392 | Actinin, alpha 1 | |
| Transport | |||
| 13.1 | 66.5 | X81627z | Lipocalin 2 |
| 12.1 | 2 | L48687 | Sodium channel, voltage-gated, type I, beta polypeptide |
| 7.2 | U04827 | Fatty acid binding protein 7, brain | |
| 3.7 | M24417 | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | |
| 3.4 | AI842825 | Glycolipid transfer protein | |
| 3.2 | NM_033444 | Chloride intracellular channel 1 | |
| 3.2 | X99347 | Lipopolysaccharide binding protein | |
| 2.9 | L13732 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | |
| 2.8 | U72680 | FXYD domain-containing ion transport regulator 5 | |
| 2.8 | X60367 | Retinol binding protein 1, cellular | |
| 2.5 | U27315 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | |
| 2.4 | AI842065 | Expressed sequence AW538430 | |
| 2.3 | AI849583 | RIKEN cDNA 6330416G13 gene | |
| 2.3 | AI852578 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 | |
| 2.1 | D87661 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | |
| 2.1 | U28960 | Phospholipid transfer protein | |
| -2 | -2.2 | AA670737 | RIKEN cDNA 1700013L23 gene |
| -2.1 | M16360 | Major urinary protein 5 | |
| -2.1 | AF072757 | Solute carrier family 27 (fatty acid transporter), member 2 | |
| -2.1 | M16358 | Major urinary protein 4 | |
| -2.1 | L28836 | ATP-binding cassette, sub-family D (ALD), member 3 | |
| -2.3 | U38652 | Solute carrier family 22 (organic cation transporter), member 1 | |
| -2.3 | M16357 | Major urinary protein 3 | |
| -2.3 | U95131 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | |
| -2.3 | -2.3 | AV355798 | Major urinary protein 2 |
| -2.3 | AV104178 | Serine (or cysteine) proteinase inhibitor, clade A, member 6 | |
| -2.4 | U95132 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | |
| -2.4 | M16359 | Major urinary protein 1 | |
| -2.4 | AB028737 | ATP-binding cassette, sub-family C (CFTR/MRP), member 6 | |
| -2.6 | -2 | D00073 | Transthyretin |
| -2.9 | AJ011080 | Afamin | |
| -3.8 | -3.8 | AI647632 | Similar to putative integral membrane transport protein |
| -4.4 | AI255271 | Major urinary protein 2 | |
| -6.4 | Y14660 | Fatty acid binding protein 1, liver | |
| -6.6 | X70533 | Serine (or cysteine) proteinase inhibitor, clade A, member 6 | |
| 4 | M55413 | Group-specific component | |
| 2.7 | AF047838 | Chloride channel calcium activated 1 | |
| 2.7 | AI849587 | Protein distantly related to the gamma subunit family | |
| 2.6 | D00466 | Apolipoprotein E | |
| 2.4 | AI661431 | Aquaporin 2 | |
| 2.3 | AI197481 | Amiloride binding protein 1 (amine oxidase, copper-containing) | |
| -2.1 | AI606956 | Solute carrier family 2 (facilitated glucose transporter), member 5 | |
| -2.1 | AW122706 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | |
| -2.2 | AI120514 | Solute carrier family 26 (sulfate transporter), member 1 | |
| -2.3 | AI837530 | Solute carrier family 9 (sodium/hydrogen exchanger), member 8 | |
| Cell surface markers and membrane proteins | |||
| 12.9 | X13333 | CD14 antigen | |
| 6.7 | X97227 | CD53 antigen | |
| 6.4 | M65027 | Glycoprotein 49 A | |
| 5.9 | D16432 | CD63 antigen | |
| 5.3 | 2.3 | AW209486 | Prostate stem cell antigen |
| 5 | 3.4 | AF024637 | TYRO protein tyrosine kinase binding protein |
| 3.6 | M58661 | CD24a antigen | |
| 3.3 | U37438 | Deleted in malignant brain tumors 1 | |
| 3.3 | M55561 | CD52 antigen | |
| 3.3 | AI854863 | RIKEN cDNA 1200015A22 gene | |
| 3 | AF039663 | Prominin 1 | |
| 2.7 | AI849180 | Integral membrane protein 2C | |
| 2.6 | AI787183 | RIKEN cDNA 0610011I04 gene | |
| 2.5 | 2.5 | X68273 | CD68 antigen |
| 2.2 | AB031386 | RIKEN cDNA 1810009M01 gene | |
| 2 | L11332 | CD38 antigen | |
| -2.5 | AI843959 | RIKEN cDNA 5730403B10 gene | |
| 3.3 | AW261569 | RIKEN cDNA D630035O19 gene | |
| 2 | AI847784 | CD34 antigen | |
| -2.5 | L23108 | CD36 antigen | |
| Transcription factors and nucleic acid binding proteins | |||
| 8.1 | 7.2 | V00727 | FBJ osteosarcoma oncogene |
| 6.2 | 3.6 | AW124113 | Brain abundant, membrane attached signal protein 1 |
| 4.9 | 2.2 | AW049031 | Core promoter element binding protein |
| 4 | M90397 | B-cell leukemia/lymphoma 3 | |
| 3.8 | M31885 | Inhibitor of DNA binding 1 | |
| 3.8 | 3 | AA614971 | Molecule possessing ankyrin-repeats induced by lipopolysaccharide |
| 3.2 | 3.6 | X61800 | CCAAT/enhancer binding protein (C/EBP), delta |
| 3.2 | AF017258 | Ribonuclease, RNase A family, 2 | |
| 2.7 | AB016424 | RNA binding motif protein 3 | |
| 2.6 | 2.4 | U19118 | Activating transcription factor 3 |
| 2.5 | AF016294 | E74-like factor 3 | |
| 2.4 | L03215 | SFFV proviral integration 1 | |
| 2.3 | AI642098 | RIKEN cDNA 4921515A04 gene | |
| 2.3 | U20735 | Jun-B oncogene | |
| 2.2 | M60523 | Inhibitor of DNA binding 3 | |
| 2.2 | D26089 | Minichromosome maintenance deficient 4 homolog (S. cerevisiae) | |
| 2.1 | 2.2 | U20344 | Kruppel-like factor 4 (gut) |
| -2 | U36799 | Retinoblastoma-like 2 | |
| -2 | AF038995 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | |
| -2.1 | L20450 | Zinc finger protein 97 | |
| -2.1 | X77602 | Upstream transcription factor 2 | |
| -2.2 | AF064088 | TGFB inducible early growth response 1 | |
| -2.2 | U95945 | One cut domain, family member 1 | |
| -2.4 | U62674 | Histone 2, H2aa1 | |
| -2.4 | AA002843 | Nuclear factor I/X | |
| -2.7 | AI834950 | Nuclear receptor subfamily 1, group D, member 1 | |
| -2.8 | AW047343 | D site albumin promoter binding protein | |
| -3.4 | X57638 | Peroxisome proliferator activated receptor alpha | |
| 4.7 | AI840339 | Ribonuclease, RNase A family 4 | |
| 2.7 | M28845 | Early growth response 1 | |
| 2.3 | X16995 | Nuclear receptor subfamily 4, group A, member 1 | |
| Metabolism | |||
| 8.5 | M13018 | Cysteine-rich protein 1 (intestinal) | |
| 6.1 | AV327760 | Stearoyl-Coenzyme A desaturase 2 | |
| 6 | 37.5 | X51547 | P lysozyme structural |
| 5.9 | AW046124 | Cytochrome b-245, alpha polypeptide | |
| 5.1 | 4.6 | M21050 | Lysozyme |
| 4.9 | X97047 | Pyruvate kinase, muscle | |
| 4.2 | AV368209 | Pyruvate kinase, muscle | |
| 4.1 | U43384 | Cytochrome b-245, beta polypeptide | |
| 4.1 | AA726364 | Lipoprotein lipase | |
| 4 | AI846517 | Cytochrome b-561 | |
| 4 | AI854821 | RIKEN cDNA 0610041P13 gene | |
| 3.8 | U13705 | Glutathione peroxidase 3 | |
| 3.8 | U12961 | NAD(P)H dehydrogenase, quinone 1 | |
| 3.6 | 2.1 | M26270 | Stearoyl-Coenzyme A desaturase 2 |
| 3.6 | M34141 | Prostaglandin-endoperoxide synthase 1 | |
| 3.6 | X07888 | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | |
| 3.5 | M31775 | Cytochrome b-245, alpha polypeptide | |
| 3.5 | AI847162 | RIKEN cDNA 1300017C10 gene | |
| 3.4 | U87147 | Flavin containing monooxygenase 3 | |
| 3.4 | AA690863 | ATPase, class VI, type 11A | |
| 3.4 | J04696 | Glutathione S-transferase, mu 2 | |
| 3.3 | X56824 | Heme oxygenase (decycling) 1 | |
| 3 | AJ238894 | Acyl-Coenzyme A thioesterase 3, mitochondrial | |
| 3 | D42048 | Squalene epoxidase | |
| 2.9 | AW060927 | Lanosterol synthase | |
| 2.8 | J03953 | Glutathione S-transferase, mu 3 | |
| 2.4 | U49350 | Cytidine 5'-triphosphate synthase | |
| 2.4 | AI594518 | Chitinase, acidic | |
| 2.2 | J02980 | Alkaline phosphatase 2, liver | |
| 2.2 | U27455 | Serine palmitoyltransferase, long chain base subunit 2 | |
| 2.2 | AI327450 | Phospholipase A2, group IB, pancreas | |
| 2.2 | AF077527 | Syndecan binding protein | |
| 2.2 | AA710635 | Colipase, pancreatic | |
| 2.1 | M62766 | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | |
| 2.1 | AW049778 | Mevalonate (diphospho) decarboxylase | |
| 2.1 | AF057368 | 7-dehydrocholesterol reductase | |
| 2 | U49385 | Cytidine 5'-triphosphate synthase 2 | |
| -2 | AW123316 | Methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | |
| -2 | AA824102 | Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | |
| -2 | AF098009 | Fatty acid amide hydrolase | |
| -2.1 | AV216468 | Expressed in non-metastatic cells 1, protein | |
| -2.1 | L42996 | Dihydrolipoamide branched chain transacylase E2 | |
| -2.1 | AI846934 | Lipin 1 | |
| -2.1 | AV071102 | Cytochrome c oxidase, subunit VIc | |
| -2.1 | AI839995 | Sarcosine dehydrogenase | |
| -2.1 | X61397 | Carbonic anhydrase 8 | |
| -2.1 | AF022894 | Sulfotransferase family 1B, member 1 | |
| -2.1 | U24493 | Tryptophan 2,3-dioxygenase | |
| -2.2 | AA675075 | Proline dehydrogenase (oxidase) 2 | |
| -2.2 | AV276715 | Aldehyde dehydrogenase family 3, subfamily A2 | |
| -2.3 | L11333 | Esterase 31 | |
| -2.3 | L11163 | Acyl-Coenzyme A dehydrogenase, short chain | |
| -2.3 | AI840013 | Peroxisomal delta3, delta2-enoyl-Coenzyme A isomerase | |
| -2.4 | M27347 | Elastase 1, pancreatic | |
| -2.4 | U32684 | Paraoxonase 1 | |
| -2.4 | M77015 | Hydroxysteroid dehydrogenase-3, delta<5>-3-beta | |
| -2.4 | AF030343 | Enoyl coenzyme A hydratase 1, peroxisomal | |
| -2.4 | AF047542 | Cytochrome P450, family 2, subfamily c, polypeptide 37 | |
| -2.4 | AF047727 | Cytochrome P450, family 2, subfamily c, polypeptide 40 | |
| -2.4 | Z14050 | Dodecenoyl-Coenzyme A delta isomerase (3,2 trans-enoyl-Coenyme A isomerase) | |
| -2.5 | D17674 | Cytochrome P450, family 2, subfamily c, polypeptide 29 | |
| -2.5 | AI844846 | 2,4-dienoyl CoA reductase 1, mitochondrial | |
| -2.6 | X83202 | Hydroxysteroid 11-beta dehydrogenase 1 | |
| -2.6 | U14390 | Aldehyde dehydrogenase family 3, subfamily A2 | |
| -2.6 | AW012588 | 3-ketoacyl-CoA thiolase B | |
| -2.7 | AI530403 | Acetyl-Coenzyme A acyltransferase 1 | |
| -2.7 | X51971 | Carbonic anhydrase 5a, mitochondrial | |
| -2.7 | AF031170 | Hydroxysteroid dehydrogenase-6, delta<5>-3-beta | |
| -2.8 | AI266885 | RIKEN cDNA 1700124F02 gene | |
| -2.9 | AF030513 | Retinol dehydrogenase 6 | |
| -3 | U15977 | Fatty acid Coenzyme A ligase, long chain 2 | |
| -3 | X04283 | Cytochrome P450, family 1, subfamily a, polypeptide 2 | |
| -3.4 | X63349 | Dopachrome tautomerase | |
| -3.6 | M15268 | Aminolevulinic acid synthase 2, erythroid | |
| -4 | D63764 | Pyruvate kinase liver and red blood cell | |
| -4.1 | AF026074 | Sulfotransferase related gene X1 | |
| -4.1 | Y14004 | Cytosolic acyl-CoA thioesterase 1 | |
| -4.3 | -3.8 | AV141027 | Cytochrome P450, family 7, subfamily b, polypeptide 1 |
| -4.3 | AJ132098 | Vanin 1 | |
| -4.6 | AW226939 | Carboxylesterase 3 | |
| -5.1 | U49861 | Deiodinase, iodothyronine, type I | |
| -6.1 | -3.4 | U36993 | Cytochrome P450, family 7, subfamily b, polypeptide 1 |
| -6.4 | U12791 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | |
| -6.6 | -2.8 | M88694 | Thioether S-methyltransferase |
| -6.9 | AF090317 | Cytochrome P450, family 8, subfamily b, polypeptide 1 | |
| -14 | AB018421 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | |
| -17.9 | Y11638 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | |
| -28 | 2.5 | AJ006474 | Carbonic anhydrase 3 |
| -37.8 | M21855 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | |
| -93.8 | L41519 | Hydroxysteroid dehydrogenase-5, delta<5>-3-beta | |
| 6.4 | AB006034 | Cytochrome P450, family 27, subfamily b, polypeptide 1 | |
| 4.8 | U49430 | Ceruloplasmin | |
| 3 | AF032466 | Arginase type II | |
| 2.9 | J05277 | Hexokinase 1 | |
| 2.6 | Z19521 | Low density lipoprotein receptor | |
| 2.6 | U04204 | Aldo-keto reductase family 1, member B8 | |
| AI848668 | Sterol-C4-methyl oxidase-like | ||
| 2.6 | U31966 | Carbonyl reductase 1 | |
| 2.5 | U49915 | Adipocyte complement related protein | |
| 2.4 | AW124337 | Microsomal glutathione S-transferase 1 | |
| 2.3 | U18975 | UDP-N-acetyl-alpha-D-galactosamine:(N-acetylneuraminyl)-galactosylglucosylceramide-beta-1,4-N-acetylgalactosaminyltransferase | |
| 2.2 | L06047 | Glutathione S-transferase, alpha 4 | |
| 2.1 | AA718169 | Resistin | |
| 2.1 | D88994 | AMP deaminase 3 | |
| 2 | AA710564 | N-acetylneuraminate pyruvate lyase | |
| -2 | U19265 | Glucosaminyl (N-acetyl) transferase 1, core 2 | |
| -2 | AB005450 | Carbonic anhydrase 14 | |
| -2.1 | M75886 | Hydroxysteroid dehydrogenase-2, delta<5>-3-beta | |
| -2.1 | AB020239 | Adenylate kinase 4 | |
| -2.2 | U48896 | UDP-glucuronosyltransferase 8 | |
| -2.2 | U89352 | Lysophospholipase 1 | |
| -2.2 | M12330 | Ornithine decarboxylase, structural | |
| -2.3 | U90535 | Flavin containing monooxygenase 5 | |
| -2.3 | AF009605 | Phosphoenolpyruvate carboxykinase 1, cytosolic | |
| -2.3 | AB015426 | Fucosyltransferase 9 | |
| -2.4 | U89906 | Alpha-methylacyl-CoA racemase | |
| -2.5 | AA840463 | Lysophospholipase 1 | |
| -3.5 | X06358 | UDP-glucuronosyltransferase 2 family, member 5 | |
| Other | |||
| 18.6 | 8.2 | U69488 | G7e protein |
| 10.9 | 2.4 | X67644 | Immediate early response 3 |
| 7.6 | U78770 | Trefoil factor 2 (spasmolytic protein 1) | |
| 2.9 | AI117936 | Mus musculus 11 days embryo head cDNA, RIKEN full-length enriched library, clone: 6230409N14 product:unknown EST, full insert sequence | |
| 2.7 | AI852545 | Transgelin 2 | |
| 2.6 | 2 | AW121336 | RIKEN cDNA 1600023A02 gene |
| 2.6 | X58196 | H19 fetal liver mRNA | |
| 2.5 | 2.2 | U25844 | Serine (or cysteine) proteinase inhibitor, clade B, member 6a |
| 2.4 | AA980164 | SPARC related modular calcium binding 2 | |
| 2.4 | D38410 | Trefoil factor 3, intestinal | |
| 2.2 | U44426 | Tumor protein D52 | |
| 2.1 | U22262 | Apolipoprotein B editing complex 1 | |
| 2.1 | 4.6 | AW230891 | Leucine-rich alpha-2-glycoprotein |
| -2.1 | U32170 | Regucalcin | |
| -2.3 | AI854813 | Mus musculus 3 days neonate thymus cDNA, RIKEN full-length enriched library, clone: A630086H07 product:RAS GTPASE-ACTIVATING-LIKE PROTEIN IQGAP2 homolog [Homo sapiens], full insert sequence | |
| -2.3 | AW049373 | RIKEN cDNA 2310016A09 gene | |
| -2.8 | AI326963 | Angiopoietin-like 4 | |
| -3 | AA797604 | Angiopoietin-like 4 | |
| -3.4 | 10.7 | AB011030 | Protein related to DAN and cerberus |
| 9.8 | AA986050 | Fibrinogen, gamma polypeptide | |
| 6.8 | M64086 | Serine (or cysteine) proteinase inhibitor, clade A, member 3N | |
| 5 | AA880891 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 | |
| 2.6 | Fibrinogen, alpha polypeptide | ||
| 2.1 | X61597 | Serine (or cysteine) proteinase inhibitor, clade A, member 3C | |
| 2 | X59520 | Cholecystokinin | |
| 2 | D13003 | Reticulocalbin | |
| -2.1 | AI314227 | RIKEN cDNA 0610006H10 gene | |
| -2.2 | AW122036 | Mus musculus transcribed sequence with strong similarity to protein ref:NP_005351.2(H.sapiens) v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian); v-maf musculoaponeurotic fibrosarcoma (avian) oncogene homolog; Avian musculoaponeurotic fibrosarcoma (MAF) protooncogene [Homo sapiens] | |
| -3.4 | M93264 | Pregnancy zone protein | |
In the kidneys of sham-operated and BDL mice, 49 and 51% of the 12 488 genes on the microarray chip were expressed, respectively. Seven days after surgery, 112 genes with GO annotation were up-regulated and 36 were down-regulated in the kidneys of BDL mice at least two-fold when compared with the sham-operated pair-fed controls. Thus the number of altered genes in kidney seven days after BDL was considerably smaller than that in liver (148 vs 378). Of the 112 GO genes up-regulated in kidney after BDL, 53 were also up-regulated in cholestatic liver. In contrast, of the 36 genes down-regulated in kidney, 7 were also down-regulated in liver (Table 1). What was particularly striking is that many of the most highly up-regulated genes in liver were also the same genes that were most highly up-regulated in kidney, irrespective of their functional class (Table 1). This suggests that both the liver and the kidney may be responding to similar transcriptional signaling molecules in this cholestatic model. For example, the acute phase gene, serum amyloid A3, was up-regulated 10.0-fold in liver and 36.1-fold in kidney, the gene encoding chemokine (C-X-C motif) ligand 1 was increased 9.6-fold in liver and 4.2-fold in kidney, and the gene encoding the transport molecule lipocalin 2 was up-regulated 13.1-fold in liver and 66.5-fold in kidney. In addition, a number of cell adhesion and extracellular matrix genes were similarly up-regulated in both liver and kidney. However, only one membrane transporter gene was up-regulated in both tissues, the gene encoding the β1 subunit of the voltage-gated sodium channel (Table 1). Interestingly, several genes for nucleic acid binding proteins were also highly up-regulated in both liver and kidney including the genes encoding the transcription factors FBJ osteosarcoma oncogene (alias c-Fos), CCAAT/enhancer binding protein (C/EBP), delta, and activating transcription factor 3.
In contrast, only seven genes were commonly down-regulated in both liver and kidney. These included the RIKEN cDNA 1700013L23 gene and the genes encoding similar to putative integral membrane transport protein, major urinary protein 2, transthyretin, cytochrome P450 7b1 (GenBank accession numbers AV141027 and U36993), and thioether S-methyltransferase.
Gene expression results from the microarray were confirmed by Northern analysis for selected genes that included cytochrome P450 7b1, organic cation transporter 1 (Oct1; solute carrier family 22, member 1) and similar to putative integral membrane transport protein from aliquots of the RNA samples utilized for the microarray (Figure 1).
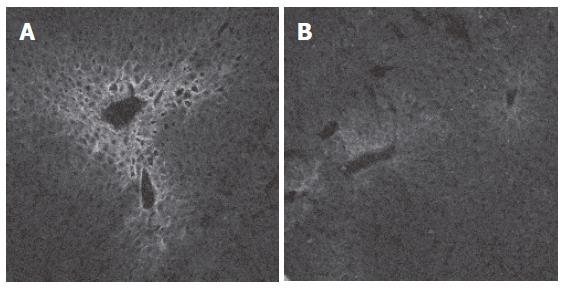
Indirect immunofluorescence was performed to illustrate the decreased expression of the organic cation transporter Oct1 in BDL mouse liver. Figures 2A and B demonstrate that the findings are consistent with the microarray and the Northern blot results and corroborate that obstructive cholestasis leads to a down-regulation of Oct1 in mouse liver similarly as demonstrated previously in rat liver following BDL[19,20].
Ligation of the common bile duct in rodents is a well-established model of obstructive cholestasis. While most previous studies have been limited to investigations of small numbers of genes and their encoded proteins, we have been able to simultaneously monitor the responses of large numbers of genes in this cholestatic model by using high-density oligonucleotide microarray technology. In contrast to a recent study which investigated gene expression in obstructive cholestasis only in the livers of BDL mice[21], we additionally monitored alterations of gene expression in the kidneys because the kidney is functionally closely linked to the liver and provides an alternative excretory route for cholephilic substances in cholestasis[5]. One of the interesting conclusions from this analysis is the finding that many of the most highly up-regulated genes were shared in both liver and kidney, possibly due to a common response to similar transcriptional signaling molecules in both tissues. The interpretation and discussion of our data is based on the assumption that changes in gene expression lead to changes in protein expression although it is known that changes at the mRNA level do not always result in changes in protein expression in certain time periods[22]. As others have done, we first evaluated the observed changes in gene expression in terms of what is already known about the effects of cholestasis. We then attempted to identify novel regulatory processes that have not yet been investigated[22].
For example, our microarray data largely confirm previous results obtained by conventional determination of transcription in obstructive cholestasis, such as the up-regulation of the canalicular cation transporter multidrug resistance P-glycoprotein 1a (Mdr1a, Abcb1a)[23] or the down-regulation of the basolateral sodium-taurocholate cotransporting polypeptide (Ntcp, Slc10a1)[24]. In addition, our gene expression profile obtained from cholestatic liver also closely matched the gene expression profile recently generated by Campbell et al[21], although there are a substantial number of additional gene alterations in our data set. This difference can be explained since Campbell et al[21] excluded genes with expression levels of less than 1 000, whereas we included genes with a mininum signal intensity of 600 and above. This approach led to the identification of a number of novel gene alterations of functional significance for the cholestatic phenotype. For instance, the decrease in expression of the gene encoding Oct1 in BDL liver in the present microarray, an alteration not reported by Campbell et al[21] but previously reported by Ogawa et al[20] in the rat, led us to study this important basolateral cationic drug transporter in more detail. We were subsequently able to demonstrate that Oct1 is indeed down-regulated in rat liver, but not in kidney, in obstructive cholestasis at the mRNA as well as the protein levels and that this decrease results in reduced hepatic uptake of the Oct1 substrate tetraethylammonium[19]. Northern analysis and immunofluorescence microscopy of hepatic Oct1 performed in the present study indicated a similar pattern in mouse and confirmed the results of our microarray.
A number of other observations emerge from this analysis that deserve further study. For example, among the cell growth-related genes, the number of genes up-regulated in liver after BDL surpassed by far the number of down-regulated genes, a pattern which might reflect the extensive fibroproliferative process and tissue remodeling that takes place in this model of obstructive cholestasis. Similarly, a large number of genes related to cell adhesion, the extracellular matrix, and the cytoskeleton were found to be altered that have not been identified yet. We presume that many of these genes may play an important but as yet to be identified role in the fibrogenic response of the liver to bile duct obstruction. Alterations in the composition of the extracellular matrix are typical features of hepatic fibrosis[13], including substantial increases of collagens and non-collagenous components[25,26]. Accordingly, we observed a uniform up-regulation of genes encoding the procollagen types Iα1, Iα2, IIIα1, IVα1, IVα2, and Vα2 in this mouse model of obstructive cholestasis. In addition, two members of the matrix metalloproteinase family, the matrix metalloproteinases 7 and 12, were up-regulated more than ten-fold following BDL when compared with the sham-operated controls. Matrix metalloproteinases represent a group of calcium-dependent enzymes involved in physiological and pathological degradation of extracellular matrix and tissue-remodeling[27]. Matrix metalloproteinase 7 (matrilysin), an enzyme which is associated with poor prognosis in hepatocellular[28] and cholangiocellular carcinomas[29], has been closely related to the fibro-proliverative process in chronic hepatitis C[30] but not in cholestatic liver diseases. In contrast, matrix metalloproteinase 12, to our knowledge, has not been associated with liver fibrosis before and deserves future attention. Interestingly, the genes encoding tissue inhibitor of metalloproteinase 1, vascular cell adhesion molecule 1 and intercellular adhesion molecule were up-regulated both in liver and kidney of BDL mice. Genes encoding the procollagen types Iα1 and IIIα1 were also increased in the kidney of BDL mice although at lower levels than in the liver. The up-regulation of fibrosis-associated factors in kidney following BDL might be due to a paracrine action of fibrogenic mediators such as connective tissue growth factor whose hepatic expression is increased in cholestasis as previously described[31,32] and confirmed in our microarray. However, the functional relevance of the increased expression of these fibrotic genes in the kidney remains to be determined. Alternatively, the simultaneous up-regulation of important regulators of transcription following BDL such as FBJ osteosarcoma oncogene, core promoter element binding protein, and activating transcription factor 3 in both liver and kidney supports the idea of coordinated gene regulation in different tissues as response to a specific stimulus. Another non-collagenous component of the extracellular matrix which was up-regulated in BDL liver is the gene for the matricellular protein secreted acidic cysteine rich glycoprotein. Matricellular proteins are a group of matrix-associated factors that mediate cell-matrix interactions but do not serve primarily as structural elements[13]. In particular, the expression of secreted acidic cysteine rich glycoprotein has been associated with cell proliferation, migration, and extracellular matrix remodeling in tissues, and secreted acidic cysteine rich glycoprotein has been found to be increased in different models of hepatic fibrosis[33].
The expression of a number of genes encoding membrane proteins and transporters that were not previously known to be affected by cholestasis was also of interest. For example, the gene encoding the β1 subunit of the voltage-gated sodium channel which is important for the maturation and function of this channel[34] was up-regulated in liver as well as in kidney of BDL mice. In contrast, the expression of the gene encoding the ATP-binding cassette transporter multidrug resistance-associated protein 6 (Mrp6, Abcc6) was reduced in cholestatic mouse liver as previously described for the rat[20]. Since mutations of human MRP6 are associated with pseudoxanthoma elasticum, a disorder characterized by calcification of the elastic fibres and abnormalities of the collagen fibrils[35], it is tempting to speculate that reduced hepatic Mrp6 expression in cholestasis might have functional implications for the development of liver fibrosis. Other genes up-regulated in cholestatic liver were the genes encoding the macrophage receptor markers CD14 antigen and CD68 antigen. Hepatic expression of both markers is increased in patients with biliary atresia[36], and expression of CD68 antigen may be an indicator of prognosis[37]. The functional significance of the concomitant CD68 antigen elevation in BDL kidney is unclear at the moment but illustrates again the close linkage between liver and kidney in this model of cholestasis and supports again a concept of coordinated gene regulation in different tissues.
In accordance with previous studies[38], obstructive cholestasis decreased the expression of a number of genes encoding cytochrome P450 isoenzymes in liver. Since BDL results in an increase in liver concentrations of bile acids[15], the down-regulation of the cytochrome P450 7b1 (oxysterol 7α-hydroxylase) and cytochrome P450 8B1 (sterol 12α-hydroxylase) genes, that encode key enzymes in the conversion of cholesterol to bile acids[39], may represent adaptive responses to minimize the liver levels of cytotoxic bile salts. The increase of the gene encoding cytochrome P450 27b1 (25-hydroxyvitamin D3 1α-hydroxylase) in BDL kidney is another interesting observation. Cytochrome P450 27b1 catalyzes the conversion of 25-hydroxyvitamin D3 to 1,25-dihydroxyvitamin D3, the last step in vitamin D activation, which takes place in kidney[40]. Thus the increase in renal cytochrome P450 27b1 expression may reflect an adaptive response to compensate for 25-hydroxyvitamin D deficiency in cholestasis. This may be a pathophysiologically important mechanism since patients with primary biliary cirrhosis often present with deficiencies of 25-hydroxyvitamin D but normal or even elevated levels of 1, 25-dihydroxyvitamin D[41].
In summary, the present study provides a comprehensive gene expression profile from mouse liver and kidney in obstructive cholestasis. Changes in gene expression were validated by Northern analysis, immunofluorescence, or comparison with the literature. The findings in this study provide new insights for generating novel hypotheses concerning the adaptive responses of gene expression in this mouse model of cholestasis.
We thank Albert Mennone and Kathy Harry (both Liver Center, Yale University School of Medicine, New Haven, CT, USA) for excellent technical assistance and Prof. Dr. Hermann Koepsell (Institut für Anatomie und Zellbiologie, Bayerische Julius-Maximilians-Universität, Würzburg, Germany) for providing the Oct1 antibody.
S- Editor Pan BR L- Editor Kumar M E- Editor Bai SH
| 1. | Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633-671. [PubMed] |
| 2. | Gartung C, Ananthanarayanan M, Rahman MA, Schuele S, Nundy S, Soroka CJ, Stolz A, Suchy FJ, Boyer JL. Down-regulation of expression and function of the rat liver Na+/bile acid cotransporter in extrahepatic cholestasis. Gastroenterology. 1996;110:199-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 168] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 3. | Dumont M, Jacquemin E, D'Hont C, Descout C, Cresteil D, Haouzi D, Desrochers M, Stieger B, Hadchouel M, Erlinger S. Expression of the liver Na+-independent organic anion transporting polypeptide (oatp-1) in rats with bile duct ligation. J Hepatol. 1997;27:1051-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Vos TA, Hooiveld GJ, Koning H, Childs S, Meijer DK, Moshage H, Jansen PL, Müller M. Up-regulation of the multidrug resistance genes, Mrp1 and Mdr1b, and down-regulation of the organic anion transporter, Mrp2, and the bile salt transporter, Spgp, in endotoxemic rat liver. Hepatology. 1998;28:1637-1644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 249] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 5. | Lee J, Azzaroli F, Wang L, Soroka CJ, Gigliozzi A, Setchell KD, Kramer W, Boyer JL. Adaptive regulation of bile salt transporters in kidney and liver in obstructive cholestasis in the rat. Gastroenterology. 2001;121:1473-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 117] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 6. | Soroka CJ, Cai SY, Boyer JL. Effects of cholestasis on the regulation of membrane transporter expression in intestine and kidney. Hepatology. 2002;36:462A. |
| 7. | Beuers U, Denk GU, Soroka CJ, Wimmer R, Rust C, Paumgartner G, Boyer JL. Taurolithocholic acid exerts cholestatic effects via phosphatidylinositol 3-kinase-dependent mechanisms in perfused rat livers and rat hepatocyte couplets. J Biol Chem. 2003;278:17810-17818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Beuers U, Probst I, Soroka C, Boyer JL, Kullak-Ublick GA, Paumgartner G. Modulation of protein kinase C by taurolithocholic acid in isolated rat hepatocytes. Hepatology. 1999;29:477-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Rust C, Gores GJ. Apoptosis and liver disease. Am J Med. 2000;108:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 132] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Patel T, Gores GJ. Apoptosis and hepatobiliary disease. Hepatology. 1995;21:1725-1741. [PubMed] |
| 11. | Oude Elferink RP, Groen AK. Mechanisms of biliary lipid secretion and their role in lipid homeostasis. Semin Liver Dis. 2000;20:293-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 12. | Bedossa P, Paradis V. Liver extracellular matrix in health and disease. J Pathol. 2003;200:504-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 190] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 13. | Schuppan D, Ruehl M, Somasundaram R, Hahn EG. Matrix as a modulator of hepatic fibrogenesis. Semin Liver Dis. 2001;21:351-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 389] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 14. | Holloway AJ, van Laar RK, Tothill RW, Bowtell DD. Options available--from start to finish--for obtaining data from DNA microarrays II. Nat Genet. 2002;32 Suppl:481-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 140] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3(Abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem. 2003;278:36688-36698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 115] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Lee JM, Trauner M, Soroka CJ, Stieger B, Meier PJ, Boyer JL. Expression of the bile salt export pump is maintained after chronic cholestasis in the rat. Gastroenterology. 2000;118:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 165] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 17. | Meyer-Wentrup F, Karbach U, Gorboulev V, Arndt P, Koepsell H. Membrane localization of the electrogenic cation transporter rOCT1 in rat liver. Biochem Biophys Res Commun. 1998;248:673-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 78] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Karbach U, Kricke J, Meyer-Wentrup F, Gorboulev V, Volk C, Loffing-Cueni D, Kaissling B, Bachmann S, Koepsell H. Localization of organic cation transporters OCT1 and OCT2 in rat kidney. Am J Physiol Renal Physiol. 2000;279:F679-F687. [PubMed] |
| 19. | Denk GU, Soroka CJ, Mennone A, Koepsell H, Beuers U, Boyer JL. Down-regulation of the organic cation transporter 1 of rat liver in obstructive cholestasis. Hepatology. 2004;39:1382-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Ogawa K, Suzuki H, Hirohashi T, Ishikawa T, Meier PJ, Hirose K, Akizawa T, Yoshioka M, Sugiyama Y. Characterization of inducible nature of MRP3 in rat liver. Am J Physiol Gastrointest Liver Physiol. 2000;278:G438-G446. [PubMed] |
| 21. | Campbell KM, Sabla GE, Bezerra JA. Transcriptional reprogramming in murine liver defines the physiologic consequences of biliary obstruction. J Hepatol. 2004;40:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Deaciuc IV, Doherty DE, Burikhanov R, Lee EY, Stromberg AJ, Peng X, de Villiers WJ. Large-scale gene profiling of the liver in a mouse model of chronic, intragastric ethanol infusion. J Hepatol. 2004;40:219-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Schrenk D, Gant TW, Preisegger KH, Silverman JA, Marino PA, Thorgeirsson SS. Induction of multidrug resistance gene expression during cholestasis in rats and nonhuman primates. Hepatology. 1993;17:854-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 59] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Zollner G, Fickert P, Silbert D, Fuchsbichler A, Stumptner C, Zatloukal K, Denk H, Trauner M. Induction of short heterodimer partner 1 precedes downregulation of Ntcp in bile duct-ligated mice. Am J Physiol Gastrointest Liver Physiol. 2002;282:G184-G191. [PubMed] |
| 25. | Rojkind M, Giambrone MA, Biempica L. Collagen types in normal and cirrhotic liver. Gastroenterology. 1979;76:710-719. [PubMed] |
| 26. | Schuppan D. Structure of the extracellular matrix in normal and fibrotic liver: collagens and glycoproteins. Semin Liver Dis. 1990;10:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 238] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 27. | Benyon RC, Arthur MJ. Extracellular matrix degradation and the role of hepatic stellate cells. Semin Liver Dis. 2001;21:373-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 370] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 28. | Yamamoto H, Itoh F, Adachi Y, Sakamoto H, Adachi M, Hinoda Y, Imai K. Relation of enhanced secretion of active matrix metalloproteinases with tumor spread in human hepatocellular carcinoma. Gastroenterology. 1997;112:1290-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Miwa S, Miyagawa S, Soeda J, Kawasaki S. Matrix metalloproteinase-7 expression and biologic aggressiveness of cholangiocellular carcinoma. Cancer. 2002;94:428-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Lichtinghagen R, Michels D, Haberkorn CI, Arndt B, Bahr M, Flemming P, Manns MP, Boeker KH. Matrix metalloproteinase (MMP)-2, MMP-7, and tissue inhibitor of metalloproteinase-1 are closely related to the fibroproliferative process in the liver during chronic hepatitis C. J Hepatol. 2001;34:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 31. | Paradis V, Dargere D, Vidaud M, De Gouville AC, Huet S, Martinez V, Gauthier JM, Ba N, Sobesky R, Ratziu V. Expression of connective tissue growth factor in experimental rat and human liver fibrosis. Hepatology. 1999;30:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 250] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 32. | Sedlaczek N, Jia JD, Bauer M, Herbst H, Ruehl M, Hahn EG, Schuppan D. Proliferating bile duct epithelial cells are a major source of connective tissue growth factor in rat biliary fibrosis. Am J Pathol. 2001;158:1239-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 116] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Frizell E, Liu SL, Abraham A, Ozaki I, Eghbali M, Sage EH, Zern MA. Expression of SPARC in normal and fibrotic livers. Hepatology. 1995;21:847-854. [PubMed] |
| 34. | Kupershmidt S, Yang T, Roden DM. Modulation of cardiac Na+ current phenotype by beta1-subunit expression. Circ Res. 1998;83:441-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 35. | Bergen AA, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H, Swart J, Kool M, van Soest S, Baas F. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet. 2000;25:228-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 421] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 36. | Tracy TF Jr, Dillon P, Fox ES, Minnick K, Vogler C. The inflammatory response in pediatric biliary disease: macrophage phenotype and distribution. J Pediatr Surg. 1996;31:121-125; discussion 125-126;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 59] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Kobayashi H, Puri P, O'Briain DS, Surana R, Miyano T. Hepatic overexpression of MHC class II antigens and macrophage-associated antigens (CD68) in patients with biliary atresia of poor prognosis. J Pediatr Surg. 1997;32:590-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Tateishi T, Watanabe M, Nakura H, Tanaka M, Kumai T, Kobayashi S. Liver damage induced by bile duct ligation affects CYP isoenzymes differently in rats. Pharmacol Toxicol. 1998;82:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 39. | Chiang JY. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev. 2002;23:443-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 352] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 40. | Omdahl JL, Bobrovnikova EV, Annalora A, Chen P, Serda R. Expression, structure-function, and molecular modeling of vitamin D P450s. J Cell Biochem. 2003;88:356-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Kaplan MM, Elta GH, Furie B, Sadowski JA, Russell RM. Fat-soluble vitamin nutriture in primary biliary cirrhosis. Gastroenterology. 1988;95:787-792. [PubMed] |