Published online Apr 14, 2006. doi: 10.3748/wjg.v12.i14.2168
Revised: May 1, 2005
Accepted: July 20, 2005
Published online: April 14, 2006
AIM: To study the role of CDH1/E-cadherin (E-cad) gene alteration profiles including mutation, loss of heterozygosity (LOH), promoter polymorphism and hypermethylation in mechanisms of CDH1 inactivation in gastric carcinoma (GC).
METHODS: Specimens were collected surgically from 70 patients with GC. Allelotyping PCR and detection of LOH, denaturing high pressure liquid chromatography and DNA sequencing, restriction fragment length polymorphism analysis, methylation specific PCR, and immunohistochemical staining were used.
RESULTS: Promoter polymorphism was not a major mechanism of E-cad inactivation. Only one truncating mutation was found in a diffuse type tumor (3%). Both LOH and promoter hypermethylation were major mechanisms of E-cad inactivation, but interestingly, there was a negative association between the fraction of allelic loss (LOH) in tumors and hypermethylation of CDH1. Therefore LOH and hypermethylation were two different tumorigenic pathways involved in GC.
CONCLUSION: Given the findings that somatic mutation was extremely low and the relationship between LOH and hypermethylation was inverse, any two combinations of these three factors cannot fulfill the classical two-hit hypothesis of CDH1 inactivation. Thus, other mechanisms operating at the transcriptional level or at the post-translational level might be required to induce E-cadherin inactivation.
- Citation: Liu YC, Shen CY, Wu HS, Hsieh TY, Chan DC, Chen CJ, Yu JC, Yu CP, Harn HJ, Chen PJ, Hsieh CB, Chen TW, Hsu HM. Mechanisms inactivating the gene for E-cadherin in sporadic gastric carcinomas. World J Gastroenterol 2006; 12(14): 2168-2173
- URL: https://www.wjgnet.com/1007-9327/full/v12/i14/2168.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i14.2168
CDH1/E-cadherin (E-cad) is a member of the family of transmembrane glycoproteins expressed on epithelial cells and is responsible for calcium-dependent cell-to-cell adhesion[1]. E-cad forms complexes and connects actin filaments with α-, β-, and γ-catenins[2,3], which are essential to neoplastic transformation and metastasis[4,5]. Loss of cell adhesion may contribute to loss of contact inhibition of growth, which is an early step in the neoplastic process. Furthermore, loss of cadherin activity may result in cancer cell detachment and metastasis[6,7].
Gastric carcinogenesis is a multi-step process with morphological progression involving multiple genetic and epigenetic events. E-cad gene (CDH1) is an important putative tumor suppressor gene. In gastric carcinomas (GCs), the reduction in E-cad expression activation of E- cad gene varies from 17% to 92%, and is more frequent in diffuse type than in intestinal type tumors[8-13]. Germline mutation of the CDH1 gene is found in all familial GCs[14,15]. Somatic mutations of CDH1 are found in more than 50% of diffuse type GCs but are not found in intestinal type GCs in Caucasians and Japanese populations[16-19]. The rate of loss of heterozygosity (LOH) ranges from 2.8% to 60% in diffuse and intestinal type tumors[16-20]. In addition to the well-known ‘two-hit’inactivation mechanism proposed by Knudson (1971), CDH1 can be silenced in GC by epigenetic promoter hypermethylation[17,21]. Besides, Li et al[22] reported that the-60C/A polymorphism has a direct effect on the transcriptional regulation of CDH1. All above previous studies of the inactivation of this gene in patients with GC have been limited in their analyses. In this study, we investigated a range of alterations in CDH1 expression profiles, including genetic mutations, LOH, promoter polymorphism, promoter hypermethylation, and immunohistochemical stain of E-cad protein together to determine possible genetic and epigenetic mechanisms of CDH1 inactivation.
Specimens were collected surgically from 70 Taiwanese patients with GC between July 1999 and July 2002 at the Division of General Surgery, Department of Surgery, Tri-Service General Hospital, Taipei, Taiwan. None of the subjects received preoperative anticancer therapy. Clinical information was obtained from medical records. Samples were taken from representative cancerous lesions and the adjacent non-cancerous epithelial parts of the tissues were flash frozen in liquid nitrogen and stored at -80°C. All tumor DNA samples were obtained by micro-dissection from 5-µm thick hematoxylin and eosin stained and paraffin embedded tissue sections[23]. Non-cancerous DNA was extracted from tissues which were flash-frozen in liquid nitrogen and stored at -80°C. All 70 samples were classified according to the Lauren’s criteria[23]: 27 were intestinal and 43 were diffuse types. The tumors were staged at the time of surgery using the standard criteria by TNM staging, with the unified international gastric cancer staging classification[24].
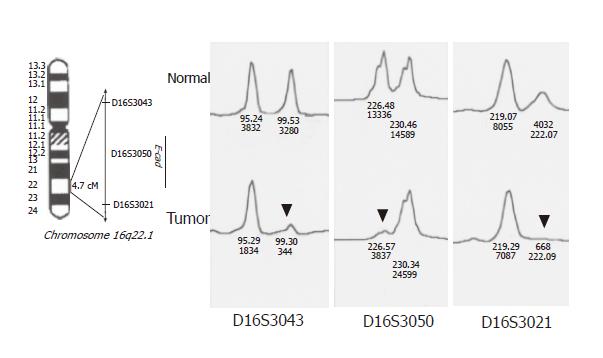
DNA samples from tumor and normal mucosal specimens were used for allelotyping PCR with fluorescent primers (markers). Three micro-satellite markers (D16S3043, D16S3050, and D16S3021) at 16q22.1 were used to detect LOH at the CDH1 locus. PCR amplification was carried out as previously described[26]. PCR products were separated electrophoretically on an ABI PRISM 377 DNA sequencer, and fluorescent signals from the differently sized alleles were recorded and analyzed using Genotyper version 2.1 and GeneScan version 3.1 software packages. A given informative marker was considered to display LOH when a threefold or greater difference was seen in the relative allele intensities of the tumor and normal DNA samples.
We used DHPLC and direct sequencing to determine inactivating mutations responsible for the loss of CDH1 expression. The promoter region and 16 exons including the exon-intron boundaries were analyzed using the previously described protocol and primer pairs[26]. The optimal conditions for DHPLC analysis of each amplicon were available as requested. All variants detected by DHPLC were re-amplified and the site of variation was identified by direct DNA sequencing using an ABI PRISM 377 DNA sequencer.
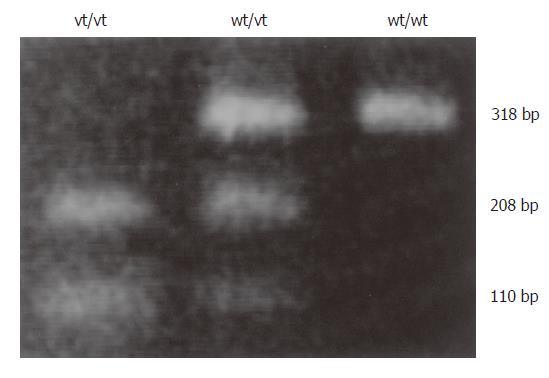
The -160 polymorphic site contained either a C or A residue. The tumor type was determined by BstEII digestion of the PCR products amplified using the primer set 5´-TGATCCCAGGTCTTAGTGAG-3´ (upstream) and 5´-AGTCTGAACTGACTT CCGCA-3´ (downstream). The 318-bp PCR product was cut into two fragments (208 and 110 bp) if it contained the A residue. To ensure that the observed polymorphism was specific and not an experimental artifact, the results were confirmed by direct DNA sequencing.
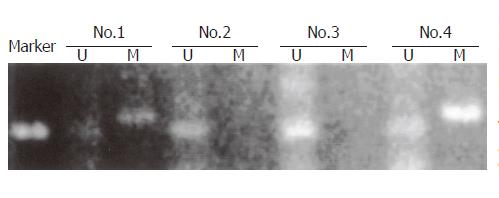
Genomic DNA was modified by bisulfite treatment, converting unmethylated cytosines to uracils and leaving methylated cytosines unchanged. MSP was performed on the treated DNA to detect all three CpG islands in the CDH1 promoter region as previously described[27]. Each unmethylated–methylated primer pair set was engineered to assess the methylation status of 4-6 CpGs with at least one CpG dinucleotide positioned at the 3´end of each primer to discriminate between methylated and unmethylated alleles following bisulfite modification. Hs578t cells, which contain a heterogeneously methylated CpG island 1 and methylated CpG islands 2 and 3, served as the positive control, and MCF7 cells were used as the negative control.
Sections (5 µm thick) were treated with monoclonal anti-E-cad antibody (Cappel, Aurora, OH, USA), then with secondary antibody. The signal was detected using a kit containing avidin–biotin complex and diaminobenzidine (DAB; Vector Laboratories, Burlingame, CA, USA). DAB produced a yellowish brown staining if the sample was positive. If more than 90% of the tumor cells exhibited intense membranous staining similar to that of normal cells, the result was considered positive (++). If the staining intensity was demonstrably reduced relative to that of normal cells and/or the staining pattern was heterogeneous (10%-90% positive), the result was deemed to be weakly positive (+). If IHC expression was completely lost or positive in less than 10% of cells, the result was defined as negative (–).
Analyses were performed using S-Plus® 2000 for Windows statistical software (CANdiensten, Amsterdam, Netherlands). Significance was assumed at P < 0.05 for all tests. Categorical variables were tested using Fisher’s exact test.
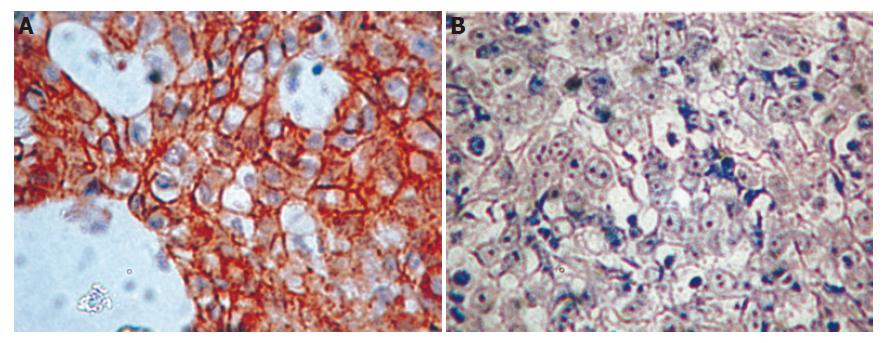
Of the 70 patients, 52 were men and 18 were women. Their median age was 69.7 years (range 32-88 years). According to Lauren’s classification, 27 and 43 tumors were intestinal and diffuse histotypes, respectively. Reduced gene expression was more frequent in diffuse type tumors (38/43, 88%) than in intestinal type tumors (13/27, 48%; P = 0.006). Representative examples of immunohistochemical staining for E-cad expression in diffuse type tumors are shown in Figure 1.
Three of the 70 patients were omitted from our analysis of the -160C/A polymorphism due to insufficient samples. Among the other 67 patients, 29 were genotype C/C (43%), 24 were genotype A/C (36%) and 14 were genotype A/A (21%) (Figure 2). There was no significant difference in the frequency of the C/A + A/A genotypes between diffuse and intestinal type tumors (27/42, 64% vs 11/25, 44%). There was no significant difference in LOH between the C/C and C/A + A/A genotypes (10/25, 40% vs 13/33, 39%). There was also no significant difference in hypermethylation between C/C and C/A + A/A genotypes (20/29, 69% vs 24/37, 65%). There was no significant difference in the frequency of the C/A + A/A genotypes between tissues with reduced and normal E-cad expression (12/17, 71% vs 27/50, 54%).
To detect allelic loss at CDH1, three micro-satellite markers (D16S3043, D16S3050, D16S3021) at 16q22.1 were used (Figure 3). The allelic status of this gene was reflected well by these three markers, because its locus was very close to the loci of these markers (LOD score > 4 estimated by linkage analysis). We considered the results for all three markers together and found heterozygosity in at least one. Of the 70 samples collected, 10 were omitted from the analysis or homozygous and could not be detected. A high frequency of allelic loss at CDH1 was detected (23/60, 38%). The frequency of LOH at CDH1 was similar between diffuse type tumors (15/38, 39%) and intestinal type tumors (8/22, 36%). Reduced E-cad expression was more frequent in LOH-positive tumors (21/23, 91%) than in LOH-negative tumors (24/37, 65%; P = 0.03).
The degree of hypermethylation estimated by MSP was defined as strongly detectable (+++, ++), detectable (+), or not detectable (–)(Figure 4). Three of the 70 samples were omitted from our analysis of hypermethylation due to insufficient samples. The CDH1 promoter was hypermethylated in 45 of these 67 GCs (67%). Hypermethylation was more frequent in diffuse type tumors (31/41, 76%) than in intestinal type tumors (13/26, 50%; P = 0.03 by Fisher’s exact test). Furthermore, hypermethylation was more frequent in GCs with reduced E-cad expression than in those with normal levels (37/45, 82% vs 12/22, 55%; P = 0.02). The fraction of allelic loss (FAL) of CDH1, calculated as the frequency of LOH at CDH1 locus, was generally inverse to the degree of hypermethylation (Tables 1, 2).
| Promoterhypermethylation | FAL | |
| Yes(++,+++) | 0.098 | |
| Yes(+) | 0.214 | |
| No | 0.377 | P = 0.03 |
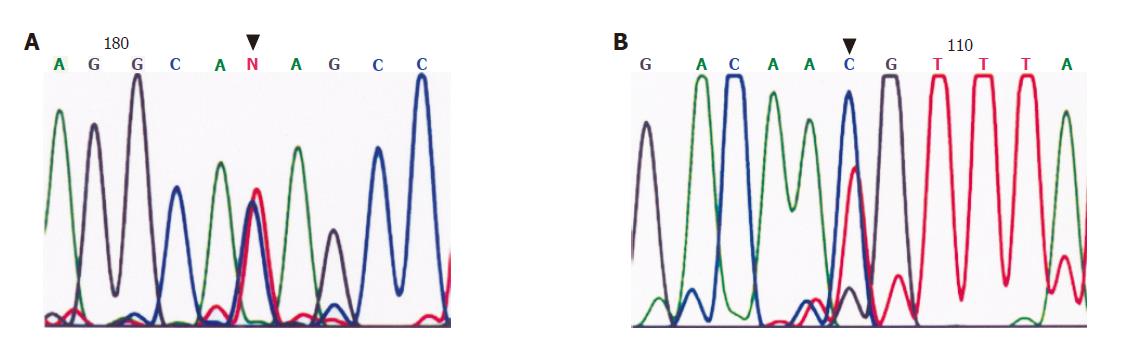
In these 70 patients, five diffuse type tumors (Case No. 15, 24, 29, 30, and 39) had a single-nucleotide polymorphism (SNP) at amino acid 692, and four diffuse type tumors (Case No. 35, 40, 59, and 63) had an SNP at position 755 . Case No.15 had a truncated mutation at position 699 (Figure 5). No CDH1 mutation was found in intestinal type tumors.
In this study, 27 and 43 tumors were of the intestinal and diffuse histotypes, respectively. Inactivation of the CDH1 gene and loss of normal E-cad expression were involved more frequently in diffuse type than in intestinal type tumors (88% vs 48%; P = 0.006). However, the percentage of reduction in E-cad expression of GC varies from 17% to 92% in previous reports[8-13]
Li et al[22] reported that the A allele of the -160C/A promoter polymorphism alters transcriptional binding, resulting in a reduction in transcriptional efficiency of 68% relative to that of the C allele. In our study, there was no significant difference in the frequency of the C/A + A/A genotypes between diffuse and intestinal type tumors. There was no significant difference in LOH and hypermethylation between the C/C and C/A + A/A genotypes. There was also no significant difference in the frequency of the C/A + A/A genotypes between tumors with reduced and normal E-cad expression, suggesting that the A allele does not play a major role in the inactivation of CDH1 and can not serve as the ‘second hit’.
Somatic mutations of CDH1 are found in more than 50% of diffuse type GCs but not in intestinal type GCs in Caucasian and Japanese populations[16-19]. A review by Berx et al[28] noted that the predominant defects in diffuse type tumors are splice mutations causing skipping in exon 8 or 9, which account for in-frame deletions, whereas mis-sense and truncating mutations are less frequent in diffuse GCs. Moreover intragenic polymorphisms arise from changes in the third (wobble) position of the respective codons and are more frequent in codons 692 and 751. In the present study, five of the diffuse type tumors had a codon 692 polymorphism and four diffuse type tumors had a codon 755 polymorphism. Only one of 38 diffuse type tumors had a truncated codon 699 mutation. Because consistent findings have been obtained by repeated detection of the same specimens, we considered this finding to be valid. Therefore, this low rate of CDH1 mutation in the Taiwanese GCs may suggest different tumorigenic mechanisms to inactivate this gene.
It was reported that the rate of LOH ranges from 2.8% to 60% in diffuse and intestinal type tumors [16-20]. A high frequency (38%) of allelic loss at CDH1 was identified in our study. The frequency of LOH was similar between the diffuse and intestinal type tumors (39% vs 36%). Reduced E-cad expression demonstrated by immunohistochemical analysis was more frequent in LOH-positive tumors than in LOH-negative tumors (91% vs 65%; P = 0.03), suggesting that LOH is a major mechanism for the inactivation of CDH1.
Tamura et al[29] and Graziano et al[30] indicated that CDH1 promoter methylation may play a major role together with mutations or deletions, in causing the inactivation of the CDH1 gene in GCs, especially in diffuse type tumors. They also reported that CDH1 promoter hypermethylation is associated with reduced E-cad expression detected immunohistochemically. In the present study, the CDH1 promoter was hypermethylated in 67% of GCs. Hypermethylation was more frequent in diffuse type tumors than in intestinal type tumors (P = 0.03). Furthermore, hypermethylation was more frequent in tumors with reduced E-cad expression than in normal E-cad expression (82% vs 55%; P = 0.02), suggesting that CDH1 promoter hypermethylation is a major mechanism for gene inactivation.
Methylation of the CDH1 promoter has been documented as the ‘second hit’ responsible for the development of hereditary diffuse GCs[31] and sporadic diffuse GCs[17] among Caucasians. Because there was only one genetic mutation in diffuse type tumors and no mutation in intestinal type tumors in this series, we examined the hypermethylated status of tumors with or without LOH at the CDH1 locus. We investigated the relationship between hypermethylation and FAL, which was estimated from the allelic status at D16S3043, D16S3050, and D16S3021. Hypermethylated tumors tended to have significantly lower FAL values (Table 1). This is contrary to the result predicted by the two-hit hypothesis. Further examination using individual markers to redefine the LOH status of tumors yielded similar results (Table 2). Therefore, cancers having lost one CDH1 allele and those carrying hypermethylated CDH1 alleles may be involved in two different tumorigenic pathways. Because the somatic mutation rate is extremely low, any two combination of these three factors cannot fulfill the classic ‘two-hit’ hypothesis. Other molecules involved in the E-cad-mediated cell-cell adhesion complex, such as the intracellular attachment proteins α, β, and γ-catenin, may be subjected to targeted inactivation[32-36]. Receptor tyrosine kinase (RTK), the main positive regulator of progression and tissue expansion, can repress E-cad function by transcriptional repression of CDH1 via the transcription factor SNAI1[37,38], posttranscriptional repression via direct or indirect phosphorylation of adheren junction components such as β-catenin[39], or RTK-associated endocytosis and degradation of the E-cad protein[40]. This more flexible status achieved either by retaining an intact allele subsequent to LOH or by regulation via epigenetic mechanisms operating at the transcriptional or posttranslational levels, could provide an advantage in counteracting the changing microenvironment during tumor progression. Further investigation is needed at the transcriptional level and the post-translational level into E-cad inactivation of GC.
| Loss of heterozygosity | |||
| Yes(%) | n(%) | Promoter hypermethylation | |
| Yes(++,+++) | 1(9.1) | 17(34.7) | |
| Yes(+) | 2(18.2) | 21(42.9) | |
| No | 8(72.7) | 11(22.5) | P = 0.001 |
In conclusion, given the finding that somatic mutation was extremely low and the relationship between LOH and hypermethylation was inverse, any two combinations of these three factors can not fulfill the classical two-hit hypothesis of E-cadherin inactivation. Thus, other mechanisms operating at the transcriptional level or at the post-translational level, might be required to inactivate E-cadherin in GC.
The authors are grateful to Miss Pei-Ei Wu and Miss Chien-Shi Wang for their technical assistance.
S- Editor Guo SY L- Editor Wang XL E- Editor Bai SH
| 1. | Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2396] [Cited by in RCA: 2422] [Article Influence: 71.2] [Reference Citation Analysis (0)] |
| 2. | Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 919] [Cited by in RCA: 936] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 3. | Grunwald GB. The structural and functional analysis of cadherin calcium-dependent cell adhesion molecules. Curr Opin Cell Biol. 1993;5:797-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 201] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Hirohashi S. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 1998;153:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 620] [Cited by in RCA: 627] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 5. | Pignatelli M, Vessey CJ. Adhesion molecules: novel molecular tools in tumor pathology. Hum Pathol. 1994;25:849-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 123] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Hashimoto M, Niwa O, Nitta Y, Takeichi M, Yokoro K. Unstable expression of E-cadherin adhesion molecules in metastatic ovarian tumor cells. Jpn J Cancer Res. 1989;80:459-463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 106] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Bussemakers MJ, van Moorselaar RJ, Giroldi LA, Ichikawa T, Isaacs JT, Takeichi M, Debruyne FM, Schalken JA. Decreased expression of E-cadherin in the progression of rat prostatic cancer. Cancer Res. 1992;52:2916-2922. [PubMed] |
| 8. | Shimoyama Y, Hirohashi S. Expression of E- and P-cadherin in gastric carcinomas. Cancer Res. 1991;51:2185-2192. [PubMed] |
| 9. | Oka H, Shiozaki H, Kobayashi K, Tahara H, Tamura S, Miyata M, Doki Y, Iihara K, Matsuyoshi N, Hirano S. Immunohistochemical evaluation of E-cadherin adhesion molecule expression in human gastric cancer. Virchows Arch A Pathol Anat Histopathol. 1992;421:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Matsuura K, Kawanishi J, Fujii S, Imamura M, Hirano S, Takeichi M, Niitsu Y. Altered expression of E-cadherin in gastric cancer tissues and carcinomatous fluid. Br J Cancer. 1992;66:1122-1130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Mayer B, Johnson JP, Leitl F, Jauch KW, Heiss MM, Schildberg FW, Birchmeier W, Funke I. E-cadherin expression in primary and metastatic gastric cancer: down-regulation correlates with cellular dedifferentiation and glandular disintegration. Cancer Res. 1993;53:1690-1695. [PubMed] |
| 12. | Gabbert H. Mechanisms of tumor invasion: evidence from in vivo observations. Cancer Metastasis Rev. 1985;4:293-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 101] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Shun CT, Wu MS, Lin JT, Wang HP, Houng RL, Lee WJ, Wang TH, Chuang SM. An immunohistochemical study of E-cadherin expression with correlations to clinicopathological features in gastric cancer. Hepatogastroenterology. 1998;45:944-949. [PubMed] |
| 14. | Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1135] [Cited by in RCA: 1140] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 15. | Gayther SA, Gorringe KL, Ramus SJ, Huntsman D, Roviello F, Grehan N, Machado JC, Pinto E, Seruca R, Halling K. Identification of germ-line E-cadherin mutations in gastric cancer families of European origin. Cancer Res. 1998;58:4086-4089. [PubMed] |
| 16. | Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Höfler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845-3852. [PubMed] |
| 17. | Machado JC, Oliveira C, Carvalho R, Soares P, Berx G, Caldas C, Seruca R, Carneiro F, Sobrinho-Simöes M. E-cadherin gene (CDH1) promoter methylation as the second hit in sporadic diffuse gastric carcinoma. Oncogene. 2001;20:1525-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 195] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 18. | Tamura G, Sakata K, Nishizuka S, Maesawa C, Suzuki Y, Iwaya T, Terashima M, Saito K, Satodate R. Inactivation of the E-cadherin gene in primary gastric carcinomas and gastric carcinoma cell lines. Jpn J Cancer Res. 1996;87:1153-1159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 82] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Ascaño JJ, Frierson H, Moskaluk CA, Harper JC, Roviello F, Jackson CE, El-Rifai W, Vindigni C, Tosi P, Powell SM. Inactivation of the E-cadherin gene in sporadic diffuse-type gastric cancer. Mod Pathol. 2001;14:942-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Machado JC, Soares P, Carneiro F, Rocha A, Beck S, Blin N, Berx G, Sobrinho-Simões M. E-cadherin gene mutations provide a genetic basis for the phenotypic divergence of mixed gastric carcinomas. Lab Invest. 1999;79:459-465. [PubMed] |
| 21. | Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL. Promoter methylation of E-cadherin gene in gastric mucosa associated with Helicobacter pylori infection and in gastric cancer. Gut. 2003;52:502-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 221] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 22. | Li LC, Chui RM, Sasaki M, Nakajima K, Perinchery G, Au HC, Nojima D, Carroll P, Dahiya R. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873-876. [PubMed] |
| 23. | Moskaluk CA, Kern SE. Microdissection and polymerase chain reaction amplification of genomic DNA from histological tissue sections. Am J Pathol. 1997;150:1547-1552. [PubMed] |
| 24. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] |
| 25. | Sobin LH, Wittekind CH (editors). International Union Against Cancers (UICC): TNM Classification of Malignant Tumors. 5th ed. New York: John Wiley 1997; PMC1716062. |
| 26. | Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919-1925. [PubMed] |
| 27. | Graff JR, Herman JG, Myöhänen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322-22329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 226] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 28. | Berx G, Becker KF, Hofler H, van Roy F. Mutations of the human E-cadherin (CDH1) gene. Hum Mutat. 1998;12:226-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 29. | Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst. 2000;92:569-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 261] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 30. | Graziano F, Arduini F, Ruzzo A, Mandolesi A, Bearzi I, Silva R, Muretto P, Testa E, Mari D, Magnani M. Combined analysis of E-cadherin gene (CDH1) promoter hypermethylation and E-cadherin protein expression in patients with gastric cancer: implications for treatment with demethylating drugs. Ann Oncol. 2004;15:489-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26:16-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 328] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 32. | Jawhari AU, Noda M, Farthing MJ, Pignatelli M. Abnormal expression and function of the E-cadherin-catenin complex in gastric carcinoma cell lines. Br J Cancer. 1999;80:322-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Ohene-Abuakwa Y, Noda M, Perenyi M, Kobayashi N, Kashima K, Hattori T, Pignatelli M. Expression of the E-cadherin/catenin (alpha-, beta-, and gamma-) complex correlates with the macroscopic appearance of early gastric cancer. J Pathol. 2000;192:433-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 34. | Yu J, Ebert MP, Miehlke S, Rost H, Lendeckel U, Leodolter A, Stolte M, Bayerdörffer E, Malfertheiner P. alpha-catenin expression is decreased in human gastric cancers and in the gastric mucosa of first degree relatives. Gut. 2000;46:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 35. | Xiangming C, Hokita S, Natsugoe S, Tanabe G, Baba M, Takao S, Kuroshima K, Aikou T. Cooccurrence of reduced expression of alpha-catenin and overexpression of p53 is a predictor of lymph node metastasis in early gastric cancer. Oncology. 1999;57:131-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Matsui S, Shiozaki H, Inoue M, Tamura S, Doki Y, Kadowaki T, Iwazawa T, Shimaya K, Nagafuchi A, Tsukita S. Immunohistochemical evaluation of alpha-catenin expression in human gastric cancer. Virchows Arch. 1994;424:375-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Batlle E, Sancho E, Francí C, Domínguez D, Monfar M, Baulida J, García De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1987] [Cited by in RCA: 2076] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 38. | Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2659] [Cited by in RCA: 2792] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 39. | Potla L, Boghaert ER, Armellino D, Frost P, Damle NK. Reduced expression of EphrinA1 (EFNA1) inhibits three-dimensional growth of HT29 colon carcinoma cells. Cancer Lett. 2002;175:187-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Kamei T, Matozaki T, Sakisaka T, Kodama A, Yokoyama S, Peng YF, Nakano K, Takaishi K, Takai Y. Coendocytosis of cadherin and c-Met coupled to disruption of cell-cell adhesion in MDCK cells--regulation by Rho, Rac and Rab small G proteins. Oncogene. 1999;18:6776-6784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 152] [Article Influence: 5.8] [Reference Citation Analysis (0)] |