Published online Mar 7, 2005. doi: 10.3748/wjg.v11.i9.1283
Revised: August 18, 2004
Accepted: September 30, 2004
Published online: March 7, 2005
AIM: Tumor endothelial markers (TEMs) are a newly discovered family of endothelial markers associated with tumor specific angiogenesis. This study sought to examine the levels of expression (qualitatively and quantitatively) for TEMs in human colon cancer.
METHODS: Human colorectal cancer tissues (n = 48) and normal background tissues (n = 31) were obtained after surgery. RNA was extracted from frozen sections for gene amplification. The expression of TEMs (TEM-1 to TEM-8) was assessed using RT-PCR and their transcript levels were determined using real-time-quantitative PCR (Q-RT-PCR).
RESULTS: TEM-1 (P = 0.01), TEM-7 (P = 0.04), TEM-7R (P = 0.03), TEM-8 (P = 0.001) significantly raised in colon cancer tissues compared with the levels detected in normal background tissues. The expressions of TEM-2 and TEM-6 were found to be not significantly different between tumor tissues and normal tissues (P>0.05). Patients who had cancer penetrating into and through the muscularis propria of the bowel wall and developed nodal involvement (Dukes C) exhibited significantly higher levels of TEM -8 compared to patients who were node negative (P<0.05). TEM-7 and TEM-7R showed high level of transcripts in Dukes C, but they were not statistically significant.
CONCLUSION: The level of the expression of TEM-1, TEM-7, TEM-7R and TEM-8 (but not TEM-2 and TEM-6) were associated with both nodal involvement and disease progression, and may therefore, have a prognostic value in colorectal cancer.
- Citation: Rmali K, Puntis M, Jiang W. Prognostic values of tumor endothelial markers in patients with colorectal cancer. World J Gastroenterol 2005; 11(9): 1283-1286
- URL: https://www.wjgnet.com/1007-9327/full/v11/i9/1283.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i9.1283
Colon cancer is the second leading cause of cancer deaths in USA and Western countries. Prognosis of patients with colorectal carcinoma is closely related to the presence of vascular and lymph node metastasis in their tumor prognosis[1-3].
Angiogenesis, defined as the sprouting of new capillaries from pre-existing vessels is characterized by expansion of the endothelium by proliferation, migration and remodeling, and is a key to cancer development and particularly metastasis[4]. Angiogenesis is a dynamic multi step process, which involves retraction of pericytes from the abluminal surface of the capillary, release of proteases from the activated endothelial cells, degradation of the ECM surrounding the pre-existing vessels, endothelial cell migration toward an angiogenic stimulus and their proliferation, formation of tube-like structures, fusion of the formed vessels and initiation of blood flow. Solid tumors are dependent on angiogenesis for growth when they reach a size of 1 or 2 mm3[5].
Despite the rapid progression in understanding the biological and the clinical significant of angiogenesis, there is very little information on markers that are specific to tumor endothelium. Most endothelial markers used in assessing angiogenesis, such as CD31, PECAM and vW factor etc., are expressed in both normal and tumor tissues.
Tumor endothelial markers (TEM1-9) were recently identified as novel endothelial cell surface markers that appear to be specific to tumor endothelial cells and are potentially involved in tumor angiogenesis[6]. TEMs are structurally and functionally conserved in mouse and human tumor endothelial cells, and are considered to be products of one of several genes elevated in human tumor endothelium and expressed at a level at least 20-fold higher in endothelial cells in vivo compared to non-endothelial cells[7]. Of particular interest, they are located on the cell-surface as they are likely to be the most accessible to pharmacological agents and may also be involved in signaling pathways that regulate angiogenesis[8].
In this study for the first time qualitatively and quantitatively, we analyzed the expression of TEMs (1, 2, 6, 7, 7R and 8) in a cohort of colorectal cancer tissues and correlated these molecules with progression of the colorectal cancer.
Colorectal tissues (n = 79) were collected from patients (with the local Research Ethic Committee approval) from patients with colorectal cancer immediately after surgical excision and stored at -20 °C until use. The samples consisted of colon tumor tissue (n = 48) together with normal background tissue from 31 of these patients, and histological information from their respective histology reports.
RNA extraction, reverse transcription Kits and PCR mix were purchased from Abgene (Surrey, UK). Total RNA was isolated using the standard guanidine isothiocyanate according to the manufacturer’s protocol as previously reported[9]. The purity and concentration of RNA was determined by spectrophotometry at 260 and 280 nm. Reverse transcription was performed and cDNA samples were synthesized in 20 µL reaction mixtures.
Conventional PCR primers were designed using the Beacon Designer software (CA) and synthesized by Life Technologies (Paisley, UK). The agarose gel extraction kit was purchased from Life Technologies. Primer sequences are given in Table 1. Conventional PCR to amplify the transcripts of TEMs (TEM1-8) was carried out using colorectal cancer and normal colorectal tissues. The reaction conditions were: 94 °C for 5 min, 36 cycles at 94 °C for 40 s, 54 °C for 30 s, 72 °C for 50 s followed by an extension phase of 10 min at 72 °C. β-actin was used as an internal housekeeping gene. The PCR products were separated on 2% and 0.8% agarose gels and stained with 10 μL ethidium bromide prior to examination and photographing under UV light.
| Primers | Sense primer (5’-3’) | Antisense primer (5’-3’) |
| TEM-1 | gtggcttcgagtgttattg | gaagagctccggatatttg |
| TEM-2 | agccatgatgaagactttgt | cttgaggtcactgttgacg |
| TEM-6 | acccgtgacgtcattttc | tgtacttgcttcgagcatc |
| TEM-7 | ggagcaggtcacgatgag | gtgaaactgcccttgtctt |
| TEM-7R | cttgattggcagtatggagt | gagatgtacatggtcccact |
| TEM-8 | catttcaagttgtcgtgaga | gacgcatattgttgttgaga |
We employed the iCycler iQ system (BioRad, Camberley, UK), to quantify the level (as copies/µL from internal standard) of TEMs in the colorectal specimens as we have previously reported[10,11]. All colorectal cDNA samples were simultaneously examined for each of the TEMs (TEM-1, -2, -6, -7, -7R and -8), along with appropriate set of plasmid standards and negative controls. Primer sets and probes used in this technique are given in Table 2.
| Molecules | Sense primer (5’-3’) | Z primer (5’-3’) |
| TEM-1 | cttgcccactgggatgat | Actgaacgtgaccgtacaacctatgaatcctctgatgg |
| TEM-2 | agtctcaccttgagtgtggt | Actgaacctgaccgtacactcctccacagcatctctta |
| TEM-6 | acccgtgaggtcattttc | Actgaacctgaccgtacattcaaccttcccatagtcag |
| TEM-7 | agaacgaccacatcacctt | Actgaacctgaccgtacatggagagagttggagtcaa |
| TEM-7R | cttgattggcagtatggagt | Actgaacctgaccgtacagtctaccgccttgagaaag |
| TEM-8 | acagggtcctctgcagctt | Actgaacctgaccgtacactttcatgccaacttgttt |
The detection of TEMs employed a universal probe system (UniPrimer™) (Intergen, Oxford, England). The UniPrimer system used two primers in conjunction with a universal probe (UniPrimer™), which recognized a specific sequence (z sequence), which had been incorporated into the primers (Table 2). A hot-start quantitation master mix (Abgene, Surrey, England) was used for the reactions.
PCR conditions for real-time QPCR were as follows: 95 °C for 12 min, followed by 50 cycles at 95 °C for 15 s, 55 °C for 60 s and 72 °C for 20 s.
Conventional RT-PCR results were analyzed by the χ2 test. Quantitative data were analyzed using Student’s t-test.
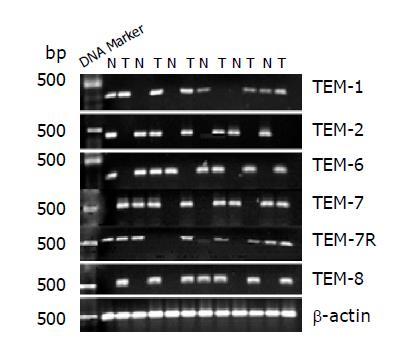
TEM-1, -7, -7R and -8 were found to be overexpressed in colon cancer tissues compared to normal tissues (P = 0.01, P = 0.04, P = 0.03 and P = 0.001, respectively). Conversely, TEM-2 and -6 expressions were found not to be different between tumor tissue and normal tissues (P = 0.61 and P = 0.56) (Figure 1, Table 3).
| Normal tissues (%) | Tumor tissues (%) | P | |
| TEM-1 | 38 | 95.5 | <0.01 |
| TEM-2 | 45 | 58.3 | >0.05 |
| TEM-6 | 35 | 56 | <0.04 |
| TEM-7 | 15 | 77.5 | <0.04 |
| TEM-7R | 12.5 | 79.5 | <0.03 |
| TEM-8 | 1 | 85 | <0.001 |
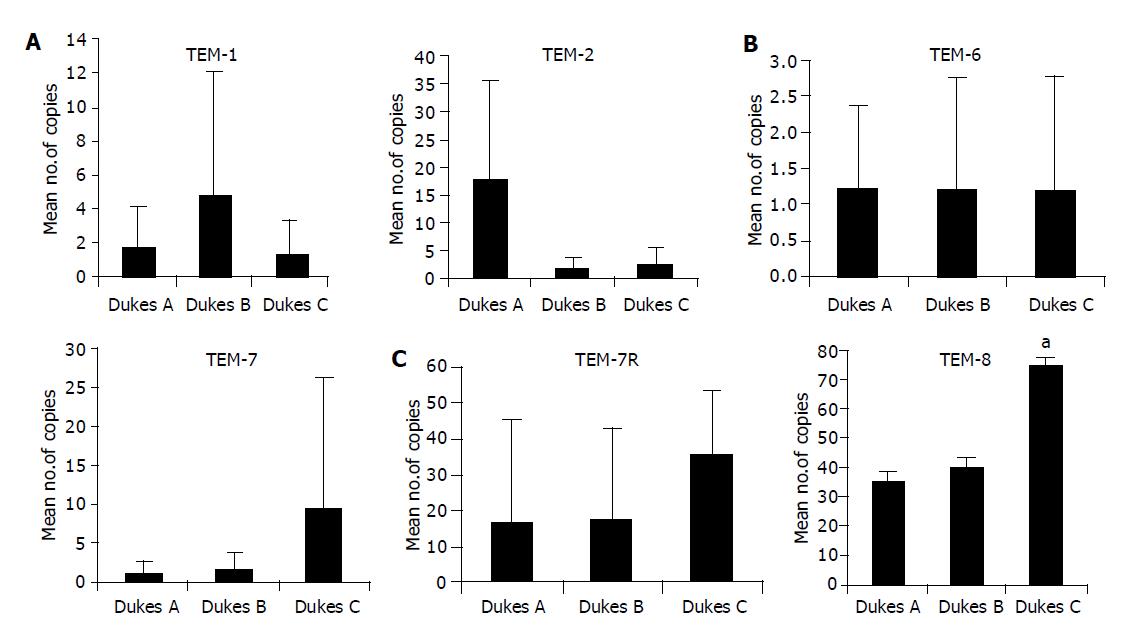
We went on to analyze, quantitatively, levels of transcript of tumor tissues in relation to Dukes staging. The number of TEM-1 transcripts was highest in Duke B, while the transcript copies of TEM-2 was significantly high in Dukes A compared to Dukes C (P<0.05). The level of expression of TEM-7 and -7R was found to be higher in Dukes C compared to Dukes A tumors; however, the difference was not statistically significant.TEM-8 expression was significantly higher in Dukes C compared to Dukes A tumors (P = 0.016) (Figure 2).
This study investigated for the first time the transcript level of a newly identified family of TEMs in colorectal cancer tissues and correlated that with tumor stage, using a quantitative approach. Previously, TEMs’ (TEM-1 and -9) expression has been found to be 10-fold higher in tumor-derived endothelium compared to endothelium derived from normal tissues[6]. These early studies have employed qualitative and semi-qualitative approach including reverse transcriptase polymerase chain reaction (RT-PCR) and in situ hybridization[7].
Our study has shown that TEMs are elevated in colorectal cancer tissues compared to normal background tissues. Although in the current study TEM-2 and -6 are expressed at a very high level in colorectal cancer tissues (Table 3); they may not be clear indicators for assessing the degree of tumor angiogenesis in colorectal cancer, since almost over half the normal tissues screened in this study were also positive for TEM-2 and -6.
On the other hand, TEM-1, -7, -7R and -8 appeared to be superior TEMs, since their expressions were significantly higher, and only a tiny proportion of normal tissues were positive. Although TEMs were initially thought to be specific only to endothelial cells in tumor tissues[6,7], our result has shown that certain TEMs do exist in normal colon mucosa, notably TEM-2 and -6. This raises some doubts as to the specificity of these markers when used in assessing tumor-induced angiogenesis. This concern was also reflected in a recent study in other tumor types, notably breast cancer[12].
The results for TEM-8 in the current study are interesting, as TEM-8 is almost absent in normal tissues but there were significantly raised levels in colon cancer tissues. The extracellular portion of TEM-8 has been shown to contain a vWF-like A domain containing a metal ion dependent adhesion site (MIDAS)[13,14]. Interestingly, the vWF-like A domain of TEM-8 has also been termed as I-domain when present within integrins and it also bears a close resemblance to a D integrin[15,16]. vWF has been shown to be an important endothelial marker in angiogenesis, whereas, integrins are cell adhesion molecules, which facilitate cell-matrix adhesion. Down-regulation in the assembly of integrins mediates adhesion complexes that have been shown to result in a gain in the invasive potential of a number of cancer cell types[17].
Our study has also shown that, TEM-8 was the only endothelial marker to be significantly elevated in colorectal cancer tissues with nodal involvement (Dukes C). Interestingly, TEM-2 and -6, which have been found highly expressed in normal colorectal tissues together with colorectal cancer, are raised in patients who had early stage cancer (Dukes A). TEM-8 appears to be unique among the cell surface TEMs in that its expression has not been detected during other forms of physiologic angiogenesis in the adult, although expression has been observed in endothelial cells of the developing mouse embryo[6,7].
This, together with previous studies strongly indicates TEM-8 as a potential marker for tumor-specific angiogenesis. In the light of the recent success of Avastin, a humanized monoclonal antibody against vascular endothelial growth factor (VEGF), in prolonging the lives of patients with metastatic colon cancer[18], it is proposed that TEM-8 may be a particularly attractive candidate for anti-angiogenic targeting colorectal cancer.
We conclude that levels of TEM-1, -2 and -7R are higher in tumors invading through the muscularis propria into subserosa, or into pericolic or perirectal tissues (Dukes A and Dukes B). TEM-8, whose expression is associated with both nodal involvement, and disease progression (Dukes C), may have significance in the progression and molecular targeting in human colorectal cancer.
Assistant Editor Guo SY Edited by Gabbe M
| 1. | Dukes CE, Bussey HJ. The spread of rectal cancer and its effect on prognosis. Br J Cancer. 1958;12:309-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 554] [Cited by in RCA: 579] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 2. | Chapuis PH, Dent OF, Fisher R, Newland RC, Pheils MT, Smyth E, Colquhoun K. A multivariate analysis of clinical and pathological variables in prognosis after resection of large bowel cancer. Br J Surg. 1985;72:698-702. [PubMed] |
| 3. | Fielding LP, Phillips RK, Fry JS, Hittinger R. Prediction of outcome after curative resection for large bowel cancer. Lancet. 1986;2:904-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 127] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2:727-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1165] [Cited by in RCA: 1079] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 5. | Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5115] [Cited by in RCA: 5897] [Article Influence: 109.2] [Reference Citation Analysis (1)] |
| 6. | St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B. Genes expressed in human tumor endothelium. Science. 2000;289:1197-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1391] [Cited by in RCA: 1377] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 7. | Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649-6655. [PubMed] |
| 8. | Nanda A, St Croix B. Tumor endothelial markers: new targets for cancer therapy. Curr Opin Oncol. 2004;16:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 95] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Rmali KA, Al-Rawi MA, Parr C, Puntis MC, Jiang WG. Upregulation of tumour endothelial marker-8 by interleukin-1beta and its impact in IL-1beta induced angiogenesis. Int J Mol Med. 2004;14:75-80. [PubMed] |
| 10. | Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, Mansel RE. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res. 2003;9:6432-6440. [PubMed] |
| 11. | Jiang WG, Douglas-Jones A, Mansel RE. Expression of peroxisome-proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int J Cancer. 2003;106:752-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 132] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Davies G, Cunnick GH, Mansel RE, Mason MD, Jiang WG. Levels of expression of endothelial markers specific to tumour-associated endothelial cells and their correlation with prognosis in patients with breast cancer. Clin Exp Metastasis. 2004;21:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Colombatti A, Bonaldo P. The superfamily of proteins with von Willebrand factor type A-like domains: one theme common to components of extracellular matrix, hemostasis, cellular adhesion, and defense mechanisms. Blood. 1991;77:2305-2315. [PubMed] |
| 14. | Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell. 1995;80:631-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 708] [Cited by in RCA: 684] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 15. | Dickeson SK, Santoro SA. Ligand recognition by the I domain-containing integrins. Cell Mol Life Sci. 1998;54:556-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 54] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Van der Vieren M, Crowe DT, Hoekstra D, Vazeux R, Hoffman PA, Grayson MH, Bochner BS, Gallatin WM, Staunton DE. The leukocyte integrin alpha D beta 2 binds VCAM-1: evidence for a binding interface between I domain and VCAM-1. J Immunol. 1999;163:1984-1990. [PubMed] |
| 17. | Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1292] [Cited by in RCA: 1281] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 18. | McCarthy M. Antiangiogenesis drug promising for metastatic colorectal cancer. Lancet. 2003;361:1959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |