Published online Feb 7, 2005. doi: 10.3748/wjg.v11.i5.629
Revised: March 14, 2004
Accepted: April 20, 2004
Published online: February 7, 2005
AIM: To understand the expression of latent and lytic genes of Epstein-Barr virus (EBV) in EBV-associated gastric carcinoma (EBVaGC) and to explore the relationship between EBV-encoded genes and development of EBVaGC at molecular level.
METHODS: One hundred and seventy-two gastric carcinoma tissues and 172 corresponding para-carcinoma tissues were tested for EBV genome by polymerase chain reaction (PCR)-Southern blotting. EBV-encoded small RNA (EBER) 1 of the PCR positive specimens was detected by in situ hybridization (ISH). Gastric carcinomas with positive EBER1 signals were classified as EBVaGCs. RT-PCR and Southern hybridization were applied to the detection of expression of nuclear antigen (EBNA) promoters (Qp, Wp and Cp), EBNA 1 and EBNA 2, latent membrane proteins (LMP) 1, 2A and 2B and lytic genes (immediate early genes BZLF1 and BRLF1, early genes BARF1 and BHRF1, late genes BcLF1 and BLLF1) in EBVaGCs.
RESULTS: Eleven EBV positive samples existed in gastric carcinoma tissues (6.39%). No EBV positive sample was found in corresponding para-carcinoma tissues. The difference between EBV positivity in carcinoma tissues and corresponding para-carcinoma tissues was significant (χ2 = 9.0909, P = 0.0026). Transcripts of Qp and EBNA1 were detected in all the 11 EBVaGCs, while both Wp and Cp were silent. EBNA2, LMP1 and LMP2B mRNA were absent in all the cases, while LMP2A mRNA was detected in 4 of the 11 cases. Of the 11 EBVaGCs, 7 exhibited BcLF1 transcripts and 2 exhibited BHRF1 transcripts. The transcripts of BZLF1 and BARF1 were detected in 5 cases, respectively. No BLLF1 and BRLF mRNA were detected.
CONCLUSION: The latent pattern of EBV in gastric carcinoma corresponds to the latency I/II. Some lytic infection genes are expressed in EBVaGCs tissues. BARF1 and BHRF1 genes may play an important role in tumorigenesis of gastric carcinoma.
- Citation: Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH, Zhao P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J Gastroenterol 2005; 11(5): 629-633
- URL: https://www.wjgnet.com/1007-9327/full/v11/i5/629.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i5.629
Epstein-Barr virus (EBV) is a tumor-related virus and EBV genome exists in many human malignant tumors, such as Burkitt’s lymphoma (BL), nasopharyngeal carcinoma (NPC), Hodgkin’s disease (HD) and B lymphocyte carcinoma in immunodeficiency patients. In recent years, EBV has also been reported to be associated with the development of gastric carcinoma. EBV has been found in most cases of rare gastric lymphoepithelioma-like carcinomas and in a small but significant proportion of common gastric adenocarcinomas. EBV-associated gastric carcinoma (EBVaGC) is observed in various histological types, such as well moderately and poorly differentiated adenocarcinomas, signet ring carcinomas[1,2]. Latent infection is a characteristic of EBV infection. It is generally thought that EBV-carrying tumors express latent infection genes but not lytic infection genes. Studies about Burkitt’s lymphoma and nasopharyngeal carcinoma (NPC) have shown that the expression of EBV genes is different in different types of malignancies and that lytic genes are also expressed[3,4]. The pathogenic role of EBV in gastric carcinomas still remains undefined. In order to identify the role of EBV in oncogenesis, the form of EBV and expression of EBV genes in tumor tissues must be understood. The aims of the present study were to understand the expression of EBV latent and lytic genes in EBVaGCs at transcriptional level, and to investigate the relationship between EBV-encoded genes and development and progress of gastric carcinomas at molecular level.
Tumor tissues and corresponding para-carcinoma tissues were dissected from the stomachs removed at surgery from 172 patients with gastric carcinoma in the Affiliated Hospital of Qingdao University Medical College, Qingdao Municipal Hospital and Yantai Yuhuangding Hospital. DNA was extracted by the standard proteinase K-sodium dodecyl sulfate (SDS) method, followed by phenol-chloroform purification. Total RNA was extracted with TRIzol reagent (Gibco BRL, Gaithersburg MD, USA) following the manufacturer’s instructions.
EBV DNA was detected by PCR and Southern hybridization analysis as previously described[5].
EBER1 of the PCR positive specimens was detected by in situ hybridization (ISH) to confirm EBV infection. ISH was carried out as previously described[6]. Briefly, paraffin-embedded sections were deparaffinized with xylene, hydrated with ethanol, and predigested with proteinase K. Then the sections were hybridized with digoxigenin (DIG)-labeled oligonucleotide probes (antisense probe: 5’-AGACACCGTCCTCACCACCCGGGACTTGTA-3’; sense probe: 5’- TCTGTGGCAGGAGTGGTG-GGCCCTGAACAT-3’) overnight at 37 °C. DIG-labeled probes were visualized by alkaline phosphatase (AP) conjugated anti-DIG antibodies. NBT/BCIP (Roche Diagnostics, Germany) was used as a substrate for AP. EBER1 sense probe was used to confirm the specificity of hybridization.
Details of the sequences and genome coordinates of primers and probes used to detect EBV transcripts are given in Table 1[1,7-11]. The probes were labeled with DIG-ddUTP by DIG oligonucleotide 3’-end labeling kit (Roche Diagnostics, Germany). Approximately 1 μg RNA of EBV-positive samples was subjected to cDNA synthesis with a reverse transcription system (Promega, USA). Three microliters of cDNA was added into a solution containing 200 μmol/L dNTPs, 500 μmol/L each primer, 1.5 mmol/L MgCl2 and 1 U Taq DNA polymerase (Promega, USA) in a total volume of 30 μL. PCR was carried out under the following conditions: first denaturation at 94 °C for 5 min, then denaturation for 45 s at 94 °C, annealing for 45 s at 55 °C, extension for 1 min at 72 °C in 35 amplification cycles, and finally extension for 5 min at 72 °C. The amplified products were electrophoresed in 2% agarose gel, transferred onto a Hybond N+ nylon membrane (Amersham Pharmacia Biotec, Ireland) and subjected to hybridization with 3’-end-DIG-labeled oligonucleotide probes. The hybridized signals were detected by alkaline phosphatase (AP) conjugated anti-DIG antibodies. The substrate of AP was CSPD (Roche Diagnostics, Germany). cDNAs from EBV-immortalized lymphoblastoid cell lines (LCL) were used as positive controls, and those from EBV-negative Ramos cells as negative controls. The integrity of RNA was checked by the parallel amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
| Transcript | Oligonucleotide sequence (5’-3’) | Product size (bp) | Genome coordinate |
| Wp | 5’primer CAGGAGATCTGGAGTCCACACAAATCCT | 131/136 | 14396-14556 |
| 5’primer GAGGAGATCTGGAGTCCACACAAATGGG | 14396-14561 | ||
| 3’primer ACTGAAGCTTGACCGGTGCCTTCTTAGGAG | 14735-14716 | ||
| probe GAGACCGAAGTGAAGGCCCTGGACCAACCC | 14561-14590 | ||
| Cp | 5’primer TGTAGATCTGATGGCATAGAGAC | 285/290 | 11342-11355 |
| 3’primer ACTGAAGCTTGACCGGTGCCTTCTTAGGAG | 14735-14716 | ||
| probe AAGGACACCGAAGACCCCCAGAG | 11356-11378 | ||
| Qp | 5’primer ATATGAGCTCGTGCGCTACCGGATGGCG | 255 | 62441-62457 |
| 3’primer GATCGAATTCCATTTCCAGGTCCTGTACCT | 107987-107967 | ||
| probe GGTGAATCTGCTCCCAGGTC | 67628-67609 | ||
| EBNA1 | 5’primer GATGAGCGTTTGGGAGAGCTGATTCTGCA | 273 | 67510-67539 |
| 3’primer TCCTCGTCCATGGTTATCAC | 108075-108056 | ||
| probe AGACCTGGGAGCAGATTCAC | 67608-67627 | ||
| EBNA2 | 5’primer GCTGCTACGCATTAGAGACC | 339 | 47892-47911 |
| 3’primer TCCTGGTAGGGATTCGAGGG | 48616-48597 | ||
| probe CAGCACTGGCGTGTGACGTGGTGTAAGTT | 48391-48420 | ||
| LMP1 | 5’primer TCCTCCTCTTGGCGCTACTG | 490 | 169383-169364 |
| 3’primer TCATCACTGTGTCGTTGTCC | 168740-168759 | ||
| probe GAACAGCACAATTCCAAGGAACAATGCCTG | 169061-169090 | ||
| LMP2A | 5’primer ATGACTCATCTCAACACATA | 280 | 166874-166893 |
| 3’primer CATGTTAGGCAAATTGCAA | 380-361 | ||
| probe ATCCAGTATGCCTGCCTGTA | 62-81 | ||
| LMP2B | 5’primer CAGTGTAATCTGCACAAAGA | 325 | 169819-169838 |
| 3’primer CATGTTAGGCAAATTGCAAA | 380-361 | ||
| probe ATCCAGTATGCCTGCCTGTA | 62-81 | ||
| BZLF1 | 5’primer ATTGCACCTTGCCGCCACCTTTG | 608 | 103194-103180 |
| 3’primer CGGCATTTTCTGGAAGCCACCCGA | 102486-102463 | ||
| probe CACTGCTGCTGCTGTTTGAACAGT | 102772-102795 | ||
| BRLF1 | 5’primer ACCATACAGGACACAACACCTC | 266 | 106166-106145 |
| 3’primer GATGTTGAGCGTGGCCATTAGC | 104959-104980 | ||
| probe GTTAGCCTCAGAAAGTCTTCCAAGCCATCC | 105140-105169 | ||
| BARF1 | 5’primer GGCTGTCACCGCTTTCTTGG | 203 | 165560-165579 |
| 3’primer AGGTGTTGGCACTTCTGTGG | 165762-165743 | ||
| probe CTGGTTTAAACTGGGCCCAGGAGAGGAGCA | 165644-165673 | ||
| BHRF1 | 5’primer GTCAAGGTTTCGTCTGTGTG | 211 | 53830-53849 |
| 3’primer TTCTCTTGCTGCTAGCTCCA | 54480-54461 | ||
| probe ATGCACACGACTGTCCCGTATACAC | 54435-54411 | ||
| BcLF1 | 5’primer TGCCCAATCCCAAGTACACGACC | 377 | 136229-136207 |
| 3’primer CAGCAGGTCATAATTGGACGGG | 135853-135874 | ||
| probe GAGAGCATTCTGTAGGTTAAACGCGAGGA | 136099-136128 | ||
| BLLF1 | 5’primer CCTACCTTGAATACAACTGG | 309 | 90860-90841 |
| 3’primer TGACGCTTGGCTGGTGGTGC | 89961-89980 | ||
| probe TGGTGACATCCGCGGTGGAT | 90731-90750 |
Software SAS 6.12 was employed to process the data with fourfold table χ2 test.
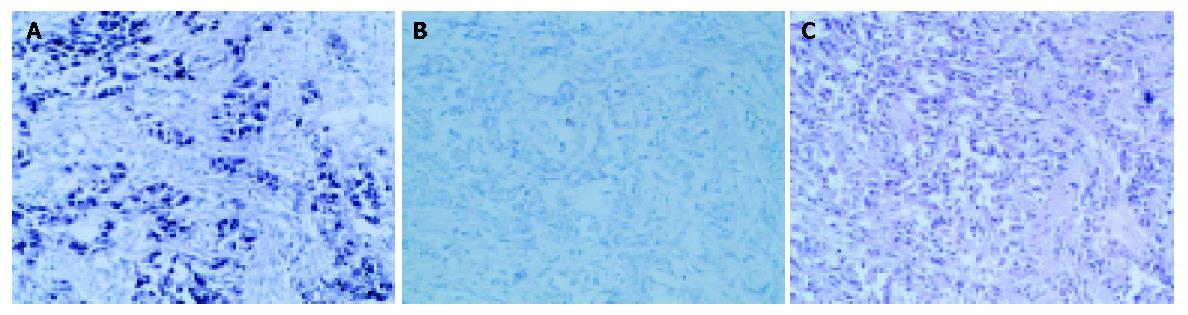
One hundred and seventy-two gastric carcinoma tissues and corresponding para-carcinoma tissues were tested for EBV genome by PCR-Southern blotting. EBER1 of the PCR positive specimens was detected by ISH to confirm EBV infection. Eleven EBV positive samples were found in gastric carcinomas (6.39%). No EBV positive sample was found in corresponding para-carcinoma tissues. The difference in EBV positivity was significant between carcinoma and corresponding para-carcinoma tissues (χ2 = 9.0909, P = 0.0026). Tumor cell nuclei of EBER1-positive cells were stained dark blue (Figure 1). Of the 11 EBVaGCs, 10 expressed EBER1 in almost all carcinoma cells, only one case expressed EBER1 in a proportion of tumor cells.
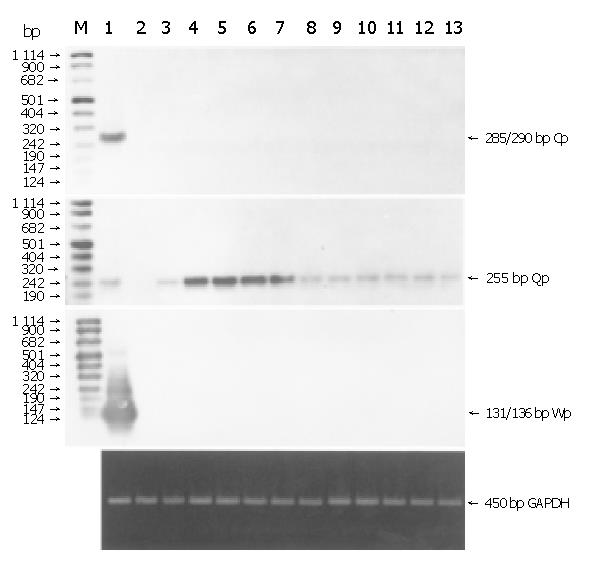
RT-PCR and Southern hybridization were performed with exon-specific primers of Qp, Wp and Cp. Transcripts of Qp were detected in all the 11 EBVaGCs, while neither Wp nor Cp transcripts were detected. The transcripts were also detected in LCL cells but not in Ramos cells (Figure 2). GAPDH mRNA was amplified to check pertinent RNA extraction. The result showed the integrity of RNA.
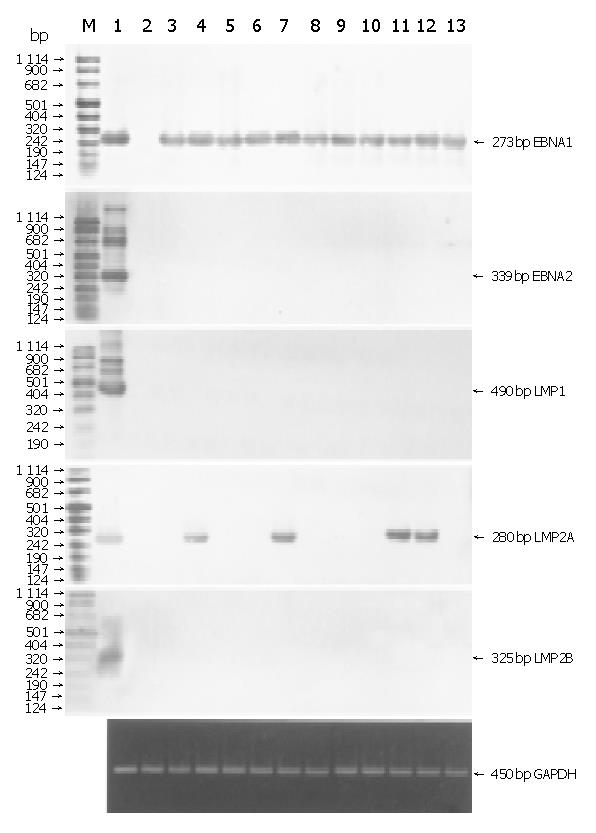
We investigated the expression of latent infection genes in 11 EBVaGCs with RT-PCR and Southern blotting (Figure 3A). EBNA1 mRNA was detected in all the cases, while EBNA2 mRNA was not detected, which was consistent with the Qp utilization for EBNA1 mRNA. LMP2A mRNA was found in 4 of the 11 cases, but neither LMP1 nor LMP2B mRNA was found in any of the cases.
To characterize the EBV lytic cycles in EBVaGCs, transcription of EBV immediate-early genes BZLF1 and BRLF1, early genes BARF1 and BHRF1 and late genes BcLF1 and BLLF1 were analyzed by RT-PCR and Southern blotting (Figure 3B). Of the 11 EBVaGCs, 7 exhibited BcLF1 transcripts and 2 exhibited BHRF1 transcripts. The transcripts of BZLF1 and BARF1 were detected respectively. No BLLF1 and BRLF mRNA were detected in the 11 EBVaGCs.
Recently, great attention has been paid to the association of EBV infection and gastric carcinoma. In the present study, 11 EBVaGCs (6.39%) were confirmed, while no EBV positive sample was found from corresponding para-carcinoma tissues (P<0.01). These results are consistent with previous reports on EBV positivity in gastric carcinoma[1,2]. Ten of the 11 EBVaGCs expressed EBER1 in almost all carcinoma cells, suggesting that EBV infection occurs early in oncogenesis with a subsequent clonal expansion of EBV-containing tumor cells as shown by other investigators using molecular genetic techniques[7,12]. One EBVaGC expressed EBER1 in a small number of gastric carcinoma cells with focal EBER1 staining, indicating that EBV infection occurs after the neoplastic transformation.
The expression of EBNA promoter genes was investigated in 11 EBVaGC tissues. Qp was clearly detected in all the cases, whereas Cp and Wp were not detected in all EBVaGCs, indicating that Qp, but not Cp or Wp, mediates EBNA transcription in EBVaGC tissues. Activation of Qp only resulted in expression of EBNA1 gene but not other EBNA genes. EBNA1 mRNA was transcribed from Qp in 11 EBVaGCs. EBNA2, LMP1 and LMP2B mRNA were not detected. Four of 11 cases exhibited LMP2A mRNA. These results are consistent with previous reports on EBVaGC[7,8]. The pattern of viral gene expression is not like the latency I of Burkitt’s lymphoma (BL) or the latency II of NPC, but corresponds to the unique latency I/II of EBV infection.
EBNA and LMP1 are essential genes for transformation of cells. Since EBNA1 is commonly expressed in 3 types of latency, it may play a similar pathogenic role in different types of tumors. Several in vitro studies have demonstrated that LMP1 can transform rodent fibroblasts and human keratinocytes, inhibit differentiation of human epithelial cells, and induce expression of epidermal growth factor receptors. These important findings strongly support that they play crucial roles in the development of non-lymphoid cell carcinomas, for example, the positivity of NPC LMP1 exceeds 80% of the NPC cases[13-15]. In the present study, EBNA1 mRNA was detected in all of the EBVaGCs, suggesting that the pathogenic role of EBNA1 is similar in EBVaGCs, NPC and BL. The absence of LMP1 expression in EBVaGCs implies that LMP1 may not be necessary for the tumors, at least not necessary for sustaining its already established malignant state. Rather, LMP1 might participate in the earlier stage of tumor development and is down-regulated thereafter. Alternatively, the lack of LMP1 may reflect the result of clonal selection of LMP1-negative tumor cells by immunological pressure because EBV-specific cytotoxic T cells are potentially directed against the viral latent membrane proteins rather than EBNA1. In fact, patients with EBVaGC normally retain virus-specific immune T-cell responses, in contrast to NPC patients[8]. It has been reported that LMP2A is involved in blocking B-cell specific signaling pathways and calcium mobilization, which might be advantageous for maintaining latent patterns of EBV infection and inhibiting EBV reactivation[16]. However, the functions of LMP2A in epithelial cells have not been analyzed yet. Serological studies have shown that NPC patients have elevated titers of antibody to both LMP2A and LMP2B, suggesting that LMP2A and LMP2B are expressed during the progression of the disease[17]. The pathogenic roles of LMP2A in the development and progression of gastric carcinoma remain to be determined.
In our study, the expression of lytic infection genes in 11 EBVaGC tissues was detected by RT-PCR analysis. We demonstrated EBV replication in part of the samples. Four cases simultaneously exhibited BZLF1, BARF1 and BcLF1 mRNA. Immediate-early genes BZLF1 and BRLF1 were necessary and sufficient to orchestrate the switch from latency to lytic replication and expression of early and late genes. BRLF1 mRNA was not detected in 11 EBVaGCs, whereas BZLF1 mRNA was detected in 5 of 11 cases. We therefore assume that BZLF1 gene activates EBV lytic replication. Early gene BHRF1 showed partial sequence homologous to human bcl-2 proto-oncogene, a gene involved in inhibiting cell apoptosis. BHRF1 protein could inhibit apoptosis of B lymphocytes and epithelial cells, and promote cell growth and transformation[18,19]. BARF1 is able to immortalize epithelial cells and fibroblast cells in vitro. Furthermore, it could activate the expression of bcl-2[1,20]. We demonstrated that 5 of 11 EBVaGCs exhibited BARF1 mRNA and 2 exhibited BHRF1 mRNA. Because EBVaGC lacks the expression of LMP1[1,7,8], BARF1 and BHRF1 genes might be the viral oncogenes in EBVaGC. Late gene BLLF1 encodes envelope glycoprotein gp320/220, which is the most abundant protein synthesized during lytic replication of EBV. The infection of B lymphocytes is mediated by adsorption of EBV gp320/220 to the receptor, CD21. In our study, BLLF1 mRNA was not found in 11 EBVaGCs. It can be proposed that EBV infects gastric epithelial cells by CD21-independent pathways.
In lytic lymphocytes, transcripts of all known EBV lytic genes have been detected. In our study, BLLF1 and BRLF1 mRNA were not detected in EBVaGC. The expression of lytic genes varied among the individual tumors analyzed. These results suggest that EBV lytic infection occurs in a small portion of EBV-infected carcinoma cells and the productive cycle is often incomplete. The same results have been reported previously[7]. Because EBER1-positive cells are detected only in carcinoma cells but not in infiltrated lymphocytes in tumor tissues, we could deny the possibility that EBV lytic infection occurs in other cells (such as infiltrated lymphocytes in tumor tissues) but not in carcinoma cells. Many studies have shown that the pattern of viral gene expression corresponds to latency I/II of EBV infection in EBVaGC. However, reports on the expression of lytic infection genes in EBVaGC are very few and have disparate results. For example, Sugiura et al[8] did not detect BZLF1 mRNA in EBVaGCs, whereas Hoshikawa et al[7] detected BZLF1, BRLF1, BLLF1 and BcLF1 mRNA in EBVaGCs.
In conclusion, the latent pattern of EBV corresponds to latency I/II and EBV lytic infection occurs in EBVaGC. BARF1 and BHRF1 may play important roles in tumorigenesis of EBVaGC. However, the mechanism by which EBV lytic infection regulates the pathogenesis and development of gastric carcinoma remains to be determined.
Assistant Editor Li WZ Edited by Zhu LH and Wang XL
| 1. | zur Hausen A, Brink AA, Craanen ME, Middeldorp JM, Meijer CJ, van den Brule AJ. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res. 2000;60:2745-2748. [PubMed] |
| 2. | Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA. 1994;91:9131-9135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 337] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 3. | Martel-Renoir D, Grunewald V, Touitou R, Schwaab G, Joab I. Qualitative analysis of the expression of Epstein-Barr virus lytic genes in nasopharyngeal carcinoma biopsies. J Gen Virol. 1995;76:1401-1408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 87] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Bashir R, Luka J, Cheloha K, Chamberlain M, Hochberg F. Expression of Epstein-Barr virus proteins in primary CNS lymphoma in AIDS patients. Neurology. 1993;43:2358-2362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Ikuta K, Satoh Y, Hoshikawa Y, Sairenji T. Detection of Epstein-Barr virus in salivas and throat washings in healthy adults and children. Microbes Infect. 2000;2:115-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 6. | Tokunaga M, Uemura Y, Tokudome T, Ishidate T, Masuda H, Okazaki E, Kaneko K, Naoe S, Ito M, Okamura A. Epstein-Barr virus related gastric cancer in Japan: a molecular patho-epidemiological study. Acta Pathol Jpn. 1993;43:574-581. [PubMed] |
| 7. | Hoshikawa Y, Satoh Y, Murakami M, Maeta M, Kaibara N, Ito H, Kurata T, Sairenji T. Evidence of lytic infection of Epstein-Barr virus (EBV) in EBV-positive gastric carcinoma. J Med Virol. 2002;66:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 8. | Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, Osato T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer. 1996;74:625-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 161] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 9. | Oudejans JJ, van den Brule AJ, Jiwa NM, de Bruin PC, Ossenkoppele GJ, van der Valk P, Walboomers JM, Meijer CJ. BHRF1, the Epstein-Barr virus (EBV) homologue of the BCL-2 protooncogene, is transcribed in EBV-associated B-cell lymphomas and in reactive lymphocytes. Blood. 1995;86:1893-1902. [PubMed] |
| 10. | Kelleher CA, Paterson RK, Dreyfus DH, Streib JE, Xu JW, Takase K, Jones JF, Gelfand EW. Epstein-Barr virus replicative gene transcription during de novo infection of human thymocytes: simultaneous early expression of BZLF-1 and its repressor RAZ. Virology. 1995;208:685-695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Prang NS, Hornef MW, Jäger M, Wagner HJ, Wolf H, Schwarzmann FM. Lytic replication of Epstein-Barr virus in the peripheral blood: analysis of viral gene expression in B lymphocytes during infectious mononucleosis and in the normal carrier state. Blood. 1997;89:1665-1677. [PubMed] |
| 12. | Gulley ML, Pulitzer DR, Eagan PA, Schneider BG. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996;27:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 124] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 636] [Cited by in RCA: 595] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 14. | Miller WE, Earp HS, Raab-Traub N. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390-4398. [PubMed] |
| 15. | Chen F, Hu LF, Ernberg I, Klein G, Winberg G. Coupled transcription of Epstein-Barr virus latent membrane protein (LMP)-1 and LMP-2B genes in nasopharyngeal carcinomas. J Gen Virol. 1995;76:131-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Miller CL, Lee JH, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci USA. 1994;91:772-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 198] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Lennette ET, Winberg G, Yadav M, Enblad G, Klein G. Antibodies to LMP2A/2B in EBV-carrying malignancies. Eur J Cancer. 1995;31A:1875-1878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Dawson CW, Dawson J, Jones R, Ward K, Young LS. Functional differences between BHRF1, the Epstein-Barr virus-encoded Bcl-2 homologue, and Bcl-2 in human epithelial cells. J Virol. 1998;72:9016-9024. [PubMed] |
| 19. | Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of the BHRF1 protein from Epstein-Barr virus, a homolog of human Bcl-2. J Mol Biol. 2003;332:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Sheng W, Decaussin G, Sumner S, Ooka T. N-terminal domain of BARF1 gene encoded by Epstein-Barr virus is essential for malignant transformation of rodent fibroblasts and activation of BCL-2. Oncogene. 2001;20:1176-1185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |