Published online Dec 28, 2005. doi: 10.3748/wjg.v11.i48.7631
Revised: June 13, 2005
Accepted: June 18, 2005
Published online: December 28, 2005
AIM: Chemokines and their receptors are crucial for immune responses in HCV and HIV infection. RANTES gene polymorphisms lead to altered gene expression and influence the natural course of HIV infection. Therefore, these mutations may also affect the course of HIV/HCV coinfection.
METHODS: We determined allele frequencies of RANTES-403 (G→A), RANTES-28 (C→G) and RANTES-IN1.1 (T→C) polymorphisms using real-time PCR and hybridization probes in patients with HIV (n = 85), HCV (n = 112), HIV/HCV coinfection (n = 121), and 109 healthy controls. Furthermore, HIV and HCV loads as well as CD4+ and CD8+ cell counts were compared between different RANTES genotypes.
RESULTS: Frequencies of RANTES-403 A, RANTES-28 G and RANTES-IN1.1 C alleles were higher in HIV infected patients than in healthy controls (-403: 28.2% vs 15.1%, P = 0.002; -28: 5.4% vs 2.8%, not significant; IN1.1: 19.0% vs 11.0%, P = 0.038). In HIV/HCV coinfected patients, these RANTES alleles were less frequent than in patients with HIV infection alone (15.4% P = 0.002; 1.7%; P = 0.048; 12.0%; not significant). Frequencies of these alleles were not significantly different between HIV/HCV positive patients, HCV positive patients and healthy controls.
CONCLUSION: All three RANTES polymorphisms showed increased frequencies of the variant allele exclusively in patients with HIV monoinfection. The finding that the frequencies of these alleles remained unaltered in HIV/HCV coinfected patients suggests that HCV coinfection interferes with selection processes associated with these alleles in HIV infection.
- Citation: Ahlenstiel G, Iwan A, Nattermann J, Bueren K, Rockstroh JK, Brackmann HH, Kupfer B, Landt O, Peled A, Sauerbruch T, Spengler U, Woitas RP. Distribution and effects of polymorphic RANTES gene alleles in HIV/HCV coinfection – A prospective cross-sectional study. World J Gastroenterol 2005; 11(48): 7631-7638
- URL: https://www.wjgnet.com/1007-9327/full/v11/i48/7631.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i48.7631
Coinfection with human immunodeficiency virus (HIV) and hepatitis C virus (HCV) is common among certain risk groups such as hemophiliacs and intravenous drug abusers (IVDA)[1]. Unfortunately, HIV/HCV coinfection is associated with an accelerated course of HCV infection leading to progressive liver disease, cirrhosis, and hepatic failure[2,3].
Chemokines and their receptors play a central role in HIV infection. During the initial steps of viral infection chemokine receptors, such as the chemokine receptor 5 (CCR5), are used as co-receptors by HIV to enter monocytes and CD4 positive T-helper cells. A mutation in the encoding region of CCR5, CCR5-△32, abrogates HIV cell entry of m-tropic HIV strains, and thus prevents HIV infection in CCR5-△32 homozygous patients[4]. Heterozygosity for the CCR5-△32 mutation is associated with delayed progression of HIV infection to AIDS[4]. Recently, we have reported that the CCR5-△32 mutation was more prevalent in hemophiliac patients with chronic hepatitis C virus infection and was associated with increased hepatitis C viral loads[5]. A study on liver biopsies of patients with chronic hepatitis C revealed that this mutation may be associated with reduced portal inflammation and fibrosis[6]. Furthermore, we have found epidemiological evidence that the CCR5-△32 mutation is a predictor of treatment failure in interferon-α monotherapy of chronic HCV infection, possibly indicating that the CCR5 receptor may also play an important role in the immune response to HCV infection[7].
The natural ligands of CCR5 are the chemokines RANTES (regulated on activation normal T cell expressed and secreted; CCL5), MIP-1α (macrophage inhibitory protein-1α; CCL3) and MIP-1β (macrophage inhibitory protein-1β; CCL4), all of which are potent inhibitors of HIV-1 cell entry[8]. Importantly, RANTES blocks the CCR5 receptor via receptor binding and down-regulation of CCR5 on T cells and macrophages. Furthermore, RANTES was reported to be critically involved in the recruitment of T cells to the liver[9]. Several single nucleotide polymorphisms in the RANTES gene have been reported to influence the natural course of HIV infection by up- or down-regulating RANTES gene activity. The most frequent of those polymorphic sites comprise RANTES-403 (G→A) and RANTES-28 (C→G) in the promoter region and RANTES-IN1.1 (T→C) in the first intron region[10,11]. Both promoter polymorphisms increase RANTES transcription and may delay HIV disease progression[11,12]. Conversely, the RANTES-IN1.1 C allele seems to decrease RANTES transcriptional activity and is probably associated with an increased risk for HIV infection and progression to AIDS[10].
Since RANTES seems to be involved in the pathogenesis of both HIV and HCV infection, we studied the effects of the RANTES gene polymorphisms in patients with HIV/HCV coinfection as compared to patients with HIV infection. To exclude that HCV infection or allele frequency in the background population contributed to our results we also included patients with HCV monoinfection as well as a group of healthy blood donors into the study.
All anti-HCV or anti-HIV positive patients of Caucasian descent attending our outpatient department between May 1999 and August 2000 were enrolled into one of the three study groups (HCV monoinfection = group I, HIV monoinfection = group II, HIV/HCV double infection = group III). None of the anti-HCV positive and anti-HIV/HCV positive patients had received interferon therapy at the time of the study. Type and duration of antiretroviral therapy and risk factors for infection were recorded in groups II and III. One hundred and nine healthy Caucasian blood donors of the Bonn University transfusion center (females 37, males 72, median age 27 years, range: 12-58 years) served as a reference group. In this reference group, HIV and HCV infection had been excluded by serology and PCR.
EDTA blood samples were obtained from each patient for genotyping of RANTES-403, -28 and -IN1.1 alleles. HCV genotype, HCV, and HIV viral loads, aminotransferase serum levels, CD4+ and CD8+ cell counts were determined in HIV, HCV, and HIV/HCV coinfected patients, respectively.
Serum aminotransferase levels were determined by routine biochemical procedures. Serologic markers of hepatitis B virus infection (HBs antigen, anti-HBs, and anti-HBc) were assessed by commercially available assays according to the manufacturer’s instructions (Abbott, Wiesbaden, Germany). CD4 and CD8 cell counts were analyzed on a FACSort™ (Becton Dickinson, Heidelberg, Germany) flow cytometer using the Simulset™ test kit (Becton Dickinson, Heidelberg, Germany). The study conformed to the ethical guidelines of the Helsinki declaration as approved by the local ethics committee.
Serum samples were analyzed for anti-HIV antibodies and p24 antigen with commercially available test kits (Abbott, Wiesbaden and Coulter, Hamburg, Germany) according to the manufacturer’s instructions. A positive ELISA result was confirmed by immunoblot (Biorad, Munich, Germany).
HIV DNA was amplified from peripheral blood leukocytes by nested PCR according to Saiki et al[13]. Amplification of the HIV-1 proviral DNA was carried out as nested PCR with the following primers for the first PCR (sense: 5-ATTTGTCATCCATCCTATTTGTTCCTGAAGGGT-3, antisense: 5-AGTGGGGGGGACATCAAGCAGCCATGCAAAT-3) and with the following primers for the second PCR (sense: 5-TGCTATGTCACTTCCCCTTGGTTCTCT-3, antisense: 5-GAGACTATCAATGAGGAAGCTGCAGAATGGGAT-3). The amplified product was detected by agarose gel electrophoresis.
HIV load was determined quantitatively using the NucliSens HIV-QT assay (Organon Teknika, Boxtel, the Netherlands). Amplified patient and calibrator RNA were quantified with different electrochemiluminescent probes in the NASBA QR system (Organon Teknika, Boxtel, the Netherlands) based on competitive internal linear standard curves[14]. This assay had a detection limit of 80 copies/mL.
HCV antibodies were detected with a microparticle enzyme immunoassay (MEIA, Axsym, Abbott, Wiesbaden, Germany). Positive results were confirmed by dot immunoassay (Matrix, Abbott, Wiesbaden, Germany). HCV RNA was detected after nucleic acid purification kit (Viral Kit, Qiagen, Hilden, Germany) followed by reverse transcription and nested polymerase chain reaction as described elsewhere[15]. The detection limit was 100 copies/mL. Quantitative determination of HCV load was done via branched DNA technology (Quantiplex HCV RNA 2.0 assay, Chiron, Emeryville, CA, USA), which has good linearity for all genotypes above its detection limit of 200 000 copies/mL serum. HCV genotypes were determined by the Innolipa II line probe assay (Innogenetics, Zwijndrecht, Belgium) according to the manufacturer’s instructions.
Genomic DNA was extracted from 200 µL EDTA-treated blood samples using the QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
Light Cycler PCR and hybridization probes: RANTES genotyping was performed using lightcycler and “fluorescence resonance energy transfer” (FRET) technology[16,17]. For amplification 2 μL light cycler-DNA master hybridization probes (Roche Diagnostics, Mannheim, Germany), 2 µL MgCl2 (25 mmol/L), 8 µL PCR-grade water were used. For genotyping of the amplified DNA, 4 pmol of each hybridization probe (TIB MolBiol, Germany) was added to the reaction mixture. Sequences of oligonucleotide primers and hybridization probes for RANTES-403 were: sense CACCTCCTTTGGGGACTGTA and antisense CCTCCGGAAATTCGAGTCTC, anchor GAGTCACTGAGTCTTCAAAGTTCCTGCTTA-F and sensor LC640-CATTACAgATCTTACCTCCTTTCCp. The sensor hybridization probe was specific for the RANTES-403G allele at a melting temperature of 62.5 °C.
Sequences of oligonucleotide primers and hybridization probes for RANTES-28 were: sense CACCTCCTTTGGGGACTgTA and antisense TGGGATGGGGTAGGCATTCTA, anchor probe GTTGCTATTTTGGAAACTCCCCTTAGG-F and sensor probe LC705-ATGCCCCTGAACTGGCCp. The sensor hybridization probe was specific for the RANTES-28G allele at a melting temperature of 60.0 °C. Sequences of oligonucleotide primers and hybridization probes for RANTES-IN1.1 were: sense CCTGGTCTTGACCACCACA and antisense GCTGACAGGCATGAGTCAGA, anchor probe LC640-CCCTCAAGGCCTACAGGTGTTCACp and sensor probe TCAGTTTTTCTGTCTTCAAGTCTAC-F. The sensor probe was specific for the C allele at a melting temperature of 65 °C.
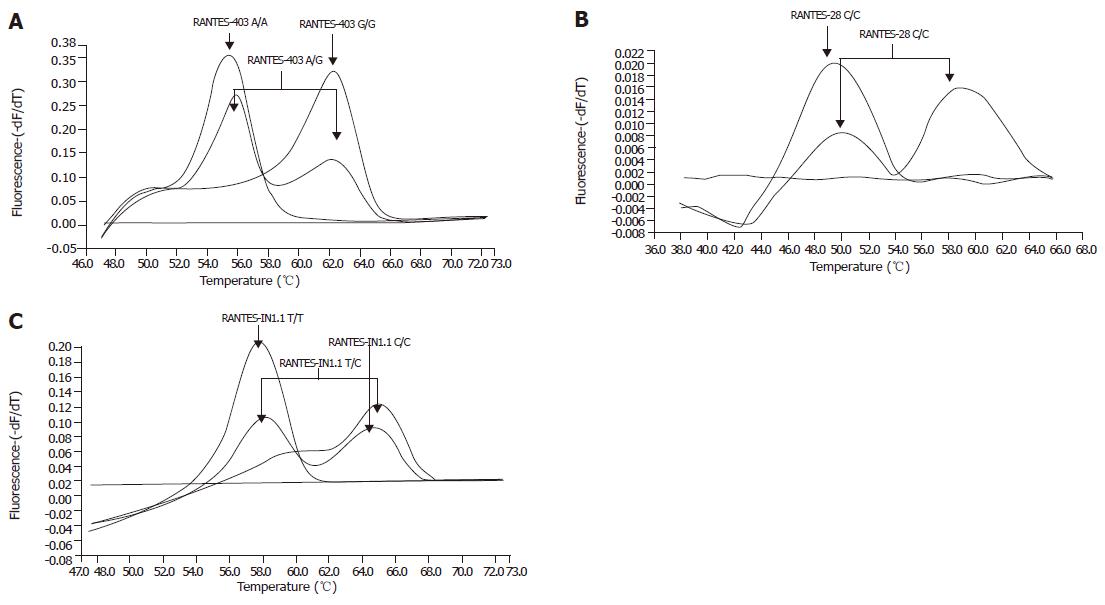
An initial denaturation step at 95 °C (60 s, ramp rate 20 °C/s) was followed by amplification for 40 cycles of denaturation (95 °C, 0 s, ramp rate 20 °C/s), annealing (63 °C, 10 s, ramp rate 20 °C/s) and extension (72 °C, 15 s, ramp rate 20 °C/s). For RANTES-IN1.1, the annealing temperature was lowered to 55 °C. After DNA amplification a melting curve was generated at 95 °C (5 s, ramp rate 20 °C/s) followed by 45 °C (15 s, ramp rate 20 °C/s) and 75 °C (0 s, ramp rate 0.1 °C/s, acquisition mode: continuous) for RANTES-403 and RANTES-IN1.1 and 95 °C, 35 °C, and 70 °C for RANTES-28. After a final cooling step for 30 s at 40 °C melting curve analysis could be performed. Representative genotyping results are given in Figures 1A-C.
RANTES genotypes and allele frequencies were compared to the healthy reference population via contingency tables using χ2 statistics and Fisher’s exact test where appropriate. Based on gene frequencies, the expected phenotype frequencies were calculated according to the Hardy–Weinberg equation and compared to the observed frequencies using χ2statistics[18]. Haplotype analysis for combined RANTES-403 and -28 genotypes as in Table 1 was performed according to Gonzalez et al[19]. In each group, different genotypes of each RANTES polymorphism were compared with respect to HIV and HCV loads, CD4+ and CD8+ cell counts using parametric (unifactorial ANOVA with Bonferroni’s correction) or non-parametric statistical analysis as appropriate (Kruskal-Wallis test followed by the Mann-Whitney test for pairwise comparison of the groups), if the number of patients with the particular genotype exceeded five. Results are given as median and range unless indicated otherwise.
| Haplotype | HIV | HIV/HCV | HCV | Controls | |
| 403 | 28 | ||||
| G | C | 71.7% | 84.0% | 80.8% | 84.2% |
| A | G | 5.4% | 1.3% | 1.8% | 2.9% |
| A | C | 22.9% | 14.2% | 17.4% | 12.9% |
| G | G | - - - | 0.5% | - - - | - - - |
| Haplotype pair (-403/-28) | HIV123 | HIV/HCV | HCV | Controls | |
| G/C | G/C | 41 (49.4%) | 82 (70.7%) | 71 (63.4%) | 73 (69.5%) |
| G/C | G/G | - - - | 1 (0.9%) | - - - | - - - |
| G/C | A/C | 28 (33.7%) | 28 (24.1%) | 37 (33.0%) | 25 (23.8%) |
| G/C | A/G | 9 (10.8%) | 2 (1.7%) | 2 (1.8%) | 6 (5.7%) |
| A/C | A/C | 5 (6.0%) | 2 (1.7%) | - - - | 1 (1.0%) |
| A/C | A/G | - - - | 1 (0.9%) | 2 (1.8%) | - - - |
In all statistical tests, P<0.05 were regarded as significant. All calculations were performed on a personal computer with SPSS 11.0 software (SPSS, Chicago, IL, USA).
One hundred and twelve anti-HCV positive, 85 anti-HIV positive, and 121 anti-HIV/HCV double-positive Caucasian patients were recruited into this study. The characteristics of these patient groups are summarized in Table 2. Hemophilia was the major risk factor for infection in HCV and HIV/HCV coinfected patients, whereas sexual transmission was the main risk factor in HIV infected patients. Rates of persistent HBs antigenemia was low in each study group (<5%), although the high prevalence of anti-HBc antibodies in the anti-HCV and the double infected groups indicated significant exposure to HBV (Table 2).
| HCV-infection(Group I) | HIV-infection(Group II) | HCV/HIV-coinfection(Group III) | Statistical significance | |
| Number | 112 | 85 | 121 | |
| Sex (Male / female) | 108 / 4 | 73 / 12 | 117 / 4 | Group I vs II P = 0.009*; group II vs III P = 0.007* |
| Age (median, range) | 39 (13 - 77) | 38 (23 - 70) | 37 (21 - 62) | |
| Risk factors (%) | ||||
| Sexual | - - - | 62 (72.9) | 1 (0.8) | Group I vs II and II vs III P < 0.001* |
| Endemic | - - - | 5 (5.9) | - - - | Group I vs II P = 0.009*; group II vs III P = 0.011* |
| I.V. drugs | - - - | 4 (4.7) | 3 (2.4) | Group I vs II P = 0.023* |
| Blood transfusion | - - - | 1 (1.2) | - - - | |
| Hemophilia | 110 (98.2) | - - - | 115 (95.0) | Group I vs II P < 0.001*; group II vs III P < 0.001* |
| Unknown | 2 (1.8) | 13 (15.3) | 2 (1.7) | Group I vs II P < 0.001*; group II vs III P < 0.001* |
| Aminotransferase activities | ||||
| ALT U/L (median, range) | 34.0 (5.2 - 226.0) | 17.0 (7.0 - 84.0) | 38.5 (1.0 - 214.0) | Group I vs II and II vs III P < 0.001 |
| AST U/L (median, range) | 19.0 (5.8 - 157.0) | 12.0 (4.3 - 106.0) | 26.1 (7.8 - 168.0) | Group I vs II P = 0.002; group I vs III P = 0.030; |
| Group II vs III P < 0.001 | ||||
| GGT U/L (median, range) | 25.2 (5.2 - 350.9) | 17.0 (6.2 - 149.0) | 42.0 (2.9 - 254.6) | Group II vs III P < 0.001 |
| HIV-status | ||||
| CD4 count (median, range) | 668 (171 - 2 039)/µL | 350 (5 - 1 142)/µL | 292.5 (6 - 1 219)/µL | Group I vs II and I vs III P < 0.001 |
| CD8 count (median, range) | 499 (62 - 1 653)/µL | 931 (88 - 2 152)/µL | 804 (39 - 3 056)/µL | Group I vs II and I vs III P < 0.001 |
| HIV load (median, range) | - - - | 300 (<80 - 830 000) | 725 (<80 - 210,000) | |
| HIV-viremia < 80 copies/mL | - - - | 47 (55.3%) | 80 (66.1%) | |
| Type of antiretroviral Therapy (%) | ||||
| Protease inhibitor-based HAART | - - - | 62 (72.9) | 58 (47.9) | Group II vs III P = 0.001 |
| NNRTI-based HAART | - - - | 11 (12.9) | 25 (20.7) | |
| Total | - - - | 73 (85.9) | 83 (68.6) | Group II vs III P = 0.007 |
| HCV-status | ||||
| HCV load (copies/mL) | 8 888 000 (n.d.-126,500,000) | - - - | 13 535 000 (n.d.-178 900 000) | Group I vs III P = 0.012 |
| HCV load (IU/mL) | 1 410 794 (n.d.-20,079,365) | - - - | 2 148 413 (n.d.-28 396 825) | |
| HCV-genotypes (%) | ||||
| Genotype 1 | 70 (62.5) | - - - | 76 (62.8) | |
| Genotype 2 | 14 (12.5) | - - - | 11 (9.1) | |
| Genotype 3 | 8 (7.1) | - - - | 21 (17.4) | Goups I vs III P = 0.028* |
| Genotype 4 | 6 (5.4) | - - - | 5 (4.1) | |
| Multiple genotypes | 1 (0.9) | - - - | 1 (0.8) | |
| Undeterminded genotype | 13 (11.6) | - - - | 7 (5.8) | |
| HBV-status (%) | ||||
| Anti-HBs+ and anti-HBc+ | 52 (46.4) | 15 (17.6) | 43 (35.5) | |
| Anti-HBc+ alone | 10 (8.9) | 5 (5.9) | 34 (28.1) | Group I vs II P < 0.001; group II vs III P = 0.008 |
| HBs-Ag+ | 1 (0.9) | 3 (3.5) | 6 (5.0) | Group I vs III P < 0.001; group II vs III P < 0.001* |
The distribution of RANTES-403, -28, and -IN1.1 alleles is shown in Table 3. The RANTES-403 A, RANTES-28 G, and RANTES-IN1.1 C alleles were more frequent in HIV monoinfected patients than in healthy controls as has been previously described[10]. The distribution of RANTES-403 genotypes in HIV infected patients differed significantly from controls (P = 0.004), whereas the RANTES-28 and RANTES-IN1.1 genotypes did not differ between the groups.
| RANTES-403 (%) | G/G | G/A | A/A | Statistics | [A]-allele frequency (%) | Statistics |
| HCV | 71 (63.4) | 39 (34.8) | 2 (1.8) | vs HIV P = 0.077 | 19.2 | vs HIV P = 0.047 |
| (n = 112) | ||||||
| HIV | 42 (49.4) | 38 (44.7) | 5 (5.9) | 28.2 | ||
| (n = 85) | ||||||
| HCV/HIV | 86 (71.7) | 31 (25.8) | 3 (2.5) | vs HIV P = 0.005 | 15.4 | vs HIV P = 0.002 |
| (n = 120) | ||||||
| Controls | 77 (70.6) | 31 (28.4) | 1 (0.9) | vs HIV P = 0.004 | 15.1 | vs HIV P = 0.002 |
| (n = 109) | ||||||
| RANTES-28 (%) | C/C | C/G | G/G | Statistics | [G]-allele frequency | Statistics |
| HCV | 108 (96.4) | 4 (3.6) | 0 (0.0) | vs HIV P = 0.078 | 1.8 | vs HIV P = 0.083 |
| (n = 112) | ||||||
| HIV | 74 (89.2) | 9 (10.8) | 0 (0.0) | 5.4 | ||
| n = 83 | ||||||
| HCV/HIV | 112 (96.6) | 4 (3.4) | 0 (0.0) | vs HIV P = 0.045 | 1.7 | vs HIV P = 0.048 |
| (n = 116) | ||||||
| Controls | 103 (94.5) | 6 (5.5) | 0 (0.0) | vs HIV P = 0.186 | 2.8 | vs HIV P = 0.195 |
| (n = 109) | ||||||
| RANTES-In1.1 (%) | T/T | T/C | C/C | Statistics | [C]-allele frequency | Statistics |
| HCV | 87 (77.7) | 25 (22.3) | 0 (0.0) | vs HIV P = 0.054 | 12.6 | vs HIV P = 0.041 |
| (n = 112) | ||||||
| HIV | 53 (63.1) | 30 (35.7) | 1 (1.2) | 19 | ||
| (n = 84) | ||||||
| HCV/HIV | 93 (76.9) | 27 (22.3) | 1 (0.8) | vs HIV P = 0.101 | 12 | vs HIV P = 0.066 |
| (n = 121) | ||||||
| Controls | 86 (78.9) | 22 (20.2) | 1 (0.9) | vs HIV P = 0.052 | 11 | vs HIV P = 0.038 |
| (n = 109) |
Unexpectedly, these higher frequencies of distinct RANTES alleles and genotypes were not observed in patients with HIV/HCV coinfection. RANTES-403 A, RANTES-28 G, and RANTES-IN1.1 C alleles were significantly less frequent than in HIV monoinfected patients (P = 0.002; P = 0.048; not significant). In contrast, the frequencies of the different RANTES alleles did not differ between HIV/HCV coinfected and HCV infected patients or healthy controls. This also held true for RANTES-403, -28, and IN1.1 genotype distribution (P = 0.005; P = 0.045; not significant). There was no deviation from the Hardy-Weinberg equilibrium in any of the groups.
Interestingly, the frequency of the combined RANTES wildtype (-403 G/G; -28 C/C; IN1.1 T/T) was lowest in HIV infected patients (49.4%) compared to healthy controls (69.9%; P = 0.007), but also compared to HCV (63.4%; not significant) and HIV/HCV coinfected patients (70.4%; P = 0.004). The [-403 G/A; -28 C/G; IN1.1 T/C] combination genotype had the highest prevalence in HIV infected patients (9.6%) differing significantly from HCV (0.9%; P = 0.005) and HIV/HCV coinfected patients (1.7%; P = 0.015) as well as controls (4.9%; not significant).
When haplotype analysis for RANTES promoter polymorphisms was performed as described previously by Gonzalez et al[19], the RANTES high producer haplotype (-403A, -28G) was most frequent in HIV monoinfected patients, whereas in HIV/HCV coinfected patients the frequency of this haplotype was similar in HCV infected patients and healthy controls (Table 3). The same finding also applies when the frequency of the (-403A, -28G) haplotype was compared between HIV and HIV/HCV infected patients (P = 0.006), between HIV and HCV infected patients (P = 0.002) and between HIV infected patients and controls (P = 0.019).
HCV and HIV viral loads as well as the numbers of CD4+ and CD8+ cell counts are given in Table 4 stratified according to RANTES genotypes. There was a trend towards higher HIV loads in HIV infected patients with the RANTES-403 G/G genotype (wildtype) compared to subjects with the G/A and A/A genotype. Furthermore, HIV infected patients with the RANTES-28 C/G genotype tended to have lower HIV loads than patients with the C/C genotype (wildtype), whereas the RANTES-IN1.1 polymorphism did not affect HIV or HCV viral loads. However, statistical analysis could not be performed for RANTES-403 A/A and RANTES-IN1.1 C/C genotypes in HCV and HCV/HIV coinfection, because these genotypes were rare in each of the groups. CD4+ and CD8+ cell counts were not affected by any of the different RANTES genotypes. Finally, HIV and HCV viral loads did not differ significantly between the different haplotypes (Figure 1).
| RANTES-403 | HCV-Infection | HIV-Infection | HIV/HCV-Infection | ||||||
| G/G(N = 71) | G/A(N = 39) | A/A(N= 2) | G/G(N= 42) | G/A(N = 38) | A/A(N = 5) | G/G(N = 86) | G/A(N = 31) | A/A(N = 3) | |
| HCV-Load | 10.8 | 4.2 | 39.0 | - - - | - - - | - - - | 13.8 | 14.0 | 1.6 |
| x 106 copies/mL | (n.d.-126.5) | (n.d.-56.6) | (n.d.-39.8) | (0.2-178.9) | (0.6-150.5) | (0.4-6.8) | |||
| HIV-Load | - - - | - - - | - - - | 455 | 380 | <80 | 550 | 770 | 9900 |
| x 103 copies/mL | (<80-830 000) | (<80-50 000) | (<80-990) | (<80-210 000) | (<80-50 000) | (2 400-27 000) | |||
| CD4 count | 683 | 621 | 916 | 353.5 | 348.5 | 300 | 302 | 279 | 293 |
| cells/µL | (171-2 039) | (282-1 544) | (787-1 045) | (51-1 111) | (5-1 142) | (28-629) | (6-1 219) | (106-651) | (149-331) |
| CD8 count | 496.5 | 499 | 581.5 | 918.5 | 927 | 1 099 | 792 | 968.5 | 675 |
| cells/µL | (62-1 653) | (185-1 183) | (425-738) | (202-1 717) | (88-2 152) | (382-1 421) | (39-2 466) | (319-3 056) | (465-895) |
| RANTES-28 | C/C (N = 108) | C/G (N = 4) | G/G (N = 0) | C/C (N = 74) | C/G (N = 9) | G/G (N = 0) | C/C (N = 112) | C/G (N = 4) | G/G (N = 0) |
| HCV-Load | 8.4 | 38.3 | - - - | - - - | - - - | - - - | 13.5 | 22.3 | - - - |
| x 106 copies/mL | (n.d.-126.5) | (29.9-39.8) | (0.2-178.9) | (0.4-45.8) | |||||
| HIV-Load | - - - | - - - | - - - | 310 | <80 | - - - | 560 | 2 150 | - - - |
| x 103 copies/mL | (<80-830 000) | (<80-3 700) | (<80-210 000) | (400-91 000) | |||||
| CD4 count | 662 | 916 | - - - | 355 | 330 | - - - | 293 | 220 | - - - |
| cells/µL | (171-2 039) | (443-1 305) | (10-1 142) | (141-637) | (13-1 219) | (149-651) | |||
| CD8 count | 499 | 581.5 | - - - | 942.5 | 636 | - - - | 828 | 602.5 | - - - |
| cells/µL | (62-1 653) | (338-739) | (202-2 152) | (362-1 997) | (39-3 056) | (397-2 401) | |||
| RANTES-IN1.1 | T/T (N = 87) | T/C (N = 25) | C/C (N = 0) | T/T (N = 53) | T/C (N = 30) | C/C (N = 1) | T/T (N = 93) | T/C (N = 27) | C/C (N = 1) |
| HCV-Load | 9.6 | 6.2 | - - - | - - - | - - - | - - - | 13.5 | 14.2 | 1.6 |
| x 106 copies/mL | (n.d.-126.5) | (0.4-56.6) | (0.2-178.9) | (0.4-150.5) | |||||
| HIV-Load | - - - | - - - | - - - | 600 | <80 | 990 | 735 | 560 | 9 900 |
| x 103 copies/mL | (<80-830 000) | (<80-38 000) | (<80-210 000) | (<80-91 000) | |||||
| CD4 count | 662 | 702 | - - - | 353 | 342 | 629 | 294 | 242 | 331 |
| cells/µL | (171-2 039) | (392-1 305) | (51-1 142) | (10-1 028) | (6-1 219) | (110-651) | |||
| CD8 count | 501 | 497 | - - - | 936.5 | 927 | 1 421 | 792 | 1 034 | 675 |
| cells/µL | (62-1 653) | (191-1 119) | (202-1 717) | (362-2 152) | (39-2 466) | (319-3 056) | |||
RANTES polymorphisms have been proposed to modify susceptibility to HIV infection and progression to AIDS[10,11,19]. In this study, the prevalence of all three polymorphic RANTES alleles (RANTES-403 A, RANTES-28 G, and RANTES-IN1.1 C) was higher in HIV monoinfected patients compared to healthy blood donors. In contrast, in HIV/HCV coinfection the frequency of the studied RANTES alleles did not differ from HCV monoinfection or healthy controls. Since we included patients with HCV monoinfection as well as a cohort of healthy blood donors into the study to exclude biased results as a consequence of HCV infection or allele frequency of the background population, the influence of RANTES alleles on HIV infection seems to be neutralized by concomitant HCV infection.
The increased frequency of distinct polymorphic RANTES alleles in HIV infection is in line with McDermott et al[20]. who observed increased susceptibility to HIV infection in patients with RANTES-403 G/A -28 C/C haplotypes. The prevalence of the RANTES-28 G allele in all groups was as low as previously described by An et al[10]. and none of our patients was homozygous for the RANTES-28 G allele[11].
The RANTES-IN1.1 C allele has been reported to be associated with an increased susceptibility for infection with HIV, which would explain the significantly increased frequency of the C allele in our HIV monoinfected patients[10]. However, none of the different RANTES alleles was significantly associated with altered HIV viral loads or CD4+ cells counts. In this context, we cannot rule out that we have missed an association between viral loads or CD4 counts and the polymorphic RANTES alleles, because our study group was rather small and not well balanced for antiviral therapy. To exclude that the presence of the CCR5-△32 allele which probably also affects susceptibility to HCV infection has influenced our findings, we tested whether any of the RANTES polymorphisms was associated or in linkage disequilibirum with CCR5-△32[5]. However, we could not find any associations between CCR5-△32 and the RANTES alleles.
The low frequency of the RANTES alleles -403A and -28G in our HCV infected patients is in accordance with Promrat et al[21]. who also could not find any association of RANTES-403 and –28 polymorphisms with susceptibility to HCV infection or laboratory parameters of chronic hepatitis C. In contrast, Hellier et al[6]. reported the RANTES-403 A/A genotype to be associated with mild portal inflammation. However, the frequency of the A/A genotype in the study of Hellier (1.9%) was not different from the frequency in our patients[6]. Therefore, it was unexpected that the prevalence of RANTES alleles known to be increased in patients with HIV monoinfection was significantly lower in our HIV/HCV coinfected patients. Additionally, prevalence of these alleles was similar to HCV monoinfected patients and controls.
Several explanations may account for these divergent results: First, none of the studies so far was controlled for differences in transmission routes. Therefore, it is unclear whether disease modifying effects of the RANTES polymorphisms are equally operative in patients with sexual transmission vs parenteral transmission.
Alternatively, our patients with HIV/HCV coinfection could represent long time survivors: The RANTES-IN1.1 C allele has been described to be associated with rapid progression to AIDS, whereas both the RANTES-403 A and the RANTES-28 G allele have been reported to delay HIV disease progression[10,11,19]. However, frequency of these polymorphic RANTES alleles did not differ from our background population. Finally, differences in genotype and allele frequencies between HIV and HIV/HCV coinfected patients might reflect differences in the physiological roles of the single polymorphisms. RANTES-403 A and RANTES-28 G alleles have been shown to up-regulate RANTES transcription, whereas RANTES-IN1.1 C has been reported to be associated with the down-regulation of RANTES promoter activity[10-12]. Thus, RANTES expression should be characterized in terms of haplotypes, defined by the combination of the various alleles, giving the possibility to explain selection effects on the course of HIV infection[10]. However, only haplotypes AC and AG were more prevalent in patients with HIV infection, whereas their frequencies in patients with HCV and HIV/HCV coinfection were identical to the healthy background population.
The selection effect of the RANTES alleles in HIV infection may be lost in patients with HCV coinfection, because interactions of HCV-specific proteins such as core and NS5A may interact with host genes to augment RANTES promotor activity[22]. Binding of the HCV-specific protein E2 to CD81 is associated with increased RANTES serum levels resulting in CCR5 internalization[23]. Therefore, HCV infection is likely to trigger increased RANTES serum levels, which in turn decrease CCR5 expression on the cell surface due to receptor internalization. This hypothesis is further supported by a recent study that shows reduced CCR1 and CCR5 surface expression on peripheral blood cells in chronic HCV infection[24]. Thus, it is intriguing to speculate that the effects of RANTES polymorphisms on RANTES expression in HIV infection are abrogated in HIV/HCV coinfection due to induction of RANTES by HCV-specific proteins. In line with this assumption, we observed reduced frequencies of patients carrying the putatively up-regulating (RANTES-403 and RANTES-28) as well as down-regulating RANTES polymorphisms (RANTES-IN1.1) in our patients with HCV and HIV/HCV coinfection.
In summary, deviations in polymorphic RANTES allele frequencies seen in HIV infection could not be confirmed in HIV/HCV coinfection. Functional studies will be required to further analyze the differences in RANTES regulation between HIV and HIV/HCV coinfection at a molecular level.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Rockstroh JK, Spengler U. HIV and hepatitis C virus co-infection. Lancet Infect Dis. 2004;4:437-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 122] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 2. | Soto B, Sánchez-Quijano A, Rodrigo L, del Olmo JA, García-Bengoechea M, Hernández-Quero J, Rey C, Abad MA, Rodríguez M, Sales Gilabert M. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 466] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Telfer P, Sabin C, Devereux H, Scott F, Dusheiko G, Lee C. The progression of HCV-associated liver disease in a cohort of haemophilic patients. Br J Haematol. 1994;87:555-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 196] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1804] [Cited by in RCA: 1735] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 5. | Woitas RP, Ahlenstiel G, Iwan A, Rockstroh JK, Brackmann HH, Kupfer B, Matz B, Offergeld R, Sauerbruch T, Spengler U. Frequency of the HIV-protective CC chemokine receptor 5-Delta32/Delta32 genotype is increased in hepatitis C. Gastroenterology. 2002;122:1721-1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Hellier S, Frodsham AJ, Hennig BJ, Klenerman P, Knapp S, Ramaley P, Satsangi J, Wright M, Zhang L, Thomas HC. Association of genetic variants of the chemokine receptor CCR5 and its ligands, RANTES and MCP-2, with outcome of HCV infection. Hepatology. 2003;38:1468-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 93] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Ahlenstiel G, Berg T, Woitas RP, Grünhage F, Iwan A, Hess L, Brackmann HH, Kupfer B, Schernick A, Sauerbruch T. Effects of the CCR5-Delta32 mutation on antiviral treatment in chronic hepatitis C. J Hepatol. 2003;39:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1667] [Cited by in RCA: 1649] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 9. | Apolinario A, Majano PL, Alvarez-Pérez E, Saez A, Lozano C, Vargas J, García-Monzón C. Increased expression of T cell chemokines and their receptors in chronic hepatitis C: relationship with the histological activity of liver disease. Am J Gastroenterol. 2002;97:2861-2870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 115] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar WL, Modi W. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci USA. 2002;99:10002-10007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 139] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Liu H, Chao D, Nakayama EE, Taguchi H, Goto M, Xin X, Takamatsu JK, Saito H, Ishikawa Y, Akaza T. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci USA. 1999;96:4581-4585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 243] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Nickel RG, Casolaro V, Wahn U, Beyer K, Barnes KC, Plunkett BS, Freidhoff LR, Sengler C, Plitt JR, Schleimer RP. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J Immunol. 2000;164:1612-1616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 188] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 13. | Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6781] [Cited by in RCA: 8412] [Article Influence: 227.4] [Reference Citation Analysis (0)] |
| 14. | van Gemen B, Kievits T, Schukkink R, van Strijp D, Malek LT, Sooknanan R, Huisman HG, Lens P. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J Virol Methods. 1993;43:177-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 107] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Imberti L, Cariani E, Bettinardi A, Zonaro A, Albertini A, Primi D. An immunoassay for specific amplified HCV sequences. J Virol Methods. 1991;34:233-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Morrison LE, Halder TC, Stols LM. Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal Biochem. 1989;183:231-244. DOI : 10.1016/0003-2697(89)90473-9. |
| 17. | Morrison LE, Stols LM. Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry. 1993;32:3095-3104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 169] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Emery AEH. An Introduction to Statistical Methods. Methodology in medical genetics. 2nd ed. London, New York: Churchill Livingstone 1986; . |
| 19. | Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, Anderson SA, Walter EA, Stephan KT, Hammer MF. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci USA. 2001;98:5199-5204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 178] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | McDermott DH, Beecroft MJ, Kleeberger CA, Al-Sharif FM, Ollier WE, Zimmerman PA, Boatin BA, Leitman SF, Detels R, Hajeer AH. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14:2671-2678 DOI : 10.1097/00002030-200012010-00006. |
| 21. | Promrat K, McDermott DH, Gonzalez CM, Kleiner DE, Koziol DE, Lessie M, Merrell M, Soza A, Heller T, Ghany M. Associations of chemokine system polymorphisms with clinical outcomes and treatment responses of chronic hepatitis C. Gastroenterology. 2003;124:352-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Soo HM, Garzino-Demo A, Hong W, Tan YH, Tan YJ, Goh PY, Lim SG, Lim SP. Expression of a full-length hepatitis C virus cDNA up-regulates the expression of CC chemokines MCP-1 and RANTES. Virology. 2002;303:253-277 DOI : 10.1006/viro.2002.1617. |
| 23. | Nattermann J, Nischalke HD, Feldmann G, Ahlenstiel G, Sauerbruch T, Spengler U. Binding of HCV E2 to CD81 induces RANTES secretion and internalization of CC chemokine receptor 5. J Viral Hepat. 2004;11:519-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Lichterfeld M, Leifeld L, Nischalke HD, Rockstroh JK, Hess L, Sauerbruch T, Spengler U. Reduced CC chemokine receptor (CCR) 1 and CCR5 surface expression on peripheral blood T lymphocytes from patients with chronic hepatitis C infection. J Infect Dis. 2002;185:1803-1807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |