Published online Dec 28, 2005. doi: 10.3748/wjg.v11.i48.7615
Revised: June 1, 2005
Accepted: June 6, 2005
Published online: December 28, 2005
AIM: To detect the common intestinal pathogenic bacteria quickly and accurately.
METHODS: A rapid (<3 h) experimental procedure was set up based upon the gene chip technology. Target genes were amplified and hybridized by oligonucleotide microarrays.
RESULTS: One hundred and seventy strains of bacteria in pure culture belonging to 11 genera were successfully discriminated under comparatively same conditions, and a series of specific hybridization maps corresponding to each kind of bacteria were obtained. When this method was applied to 26 divided cultures, 25 (96.2%) were identified.
CONCLUSION: Salmonella sp., Escherichia coli, Shigella sp., Listeria monocytogenes, Vibrio parahaemolyticus, Staphylococcus aureus, Proteus sp., Bacillus cereus, Vibrio cholerae, Enterococcus faecalis, Yersinia enterocolitica, and Campylobacter jejuni can be detected and identified by our microarrays. The accuracy, range, and discrimination power of this assay can be continually improved by adding further oligonucleotides to the arrays without any significant increase of complexity or cost.
- Citation: Jin LQ, Li JW, Wang SQ, Chao FH, Wang XW, Yuan ZQ. Detection and identification of intestinal pathogenic bacteria by hybridization to oligonucleotide microarrays. World J Gastroenterol 2005; 11(48): 7615-7619
- URL: https://www.wjgnet.com/1007-9327/full/v11/i48/7615.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i48.7615
Intestinal pathogenic bacteria exert a great threat upon human health. It is still a challenge to detect and identify bacterial pathogens quickly and accurately from the samples. Since intestinal bacterial pathogens involve a wide range of genera and species, few existing methods can meet the requirement of quick and parallel detection of these bacterial pathogens. Classical diagnostic methods, including culture and biochemical identification, immunological assay, nucleotide probe hybridization, and PCR amplification, share a common shortcoming: only one or few kinds of bacteria can be identified in a complete cycle of experiment. These serial procedures are hard to use for quick and simultaneous detection of multiple pathogenic bacteria. To meet the demands of rapid and parallel detection and identification of many common pathogenic bacteria in one experiment, we present here a new approach based on the epoch-making gene-chip (microarray) technology.
Gene chip technology is based upon the reversed solid hybridization of oligonucleotides[1,2]. The major advantages of gene chip technology, including miniature, high performance, parallelism, automation, have expanded its application in this decade[3]. Since the efficacy of gene chip technology depends heavily upon the oligonucleotide probes, careful selection of target genes and wise design of oligonucleotide probes with variable kind, sequence and amount, are cardinal factors for a good gene chip. The target genes may be species-specific. For example, the pathogenic genes[4] can be easily identified by simple PCR. However, it is impractical to use different primers for different species in gene chip technology, especially in the case where a specimen of one or more possible bacteria is given. Either a complex PCR with a mixture of many primers or a series of PCRs performed in parallel or sequential are necessary to amplify the target genes. However, the time, complexity and expense of experiment will also increase. On the contrary, if a consensus gene among many pathogenic bacteria is chosen, a single pair of carefully designed universal primers may be used to amplify the conserved stretches of DNA, which are then detected and identified by the wisely designed oligonucleotide probes. The conserved consensus genes usually chosen by many researchers are 16S ribosomal DNA (rDNA), 23S rDNA[5,6], 16S-23S rDNA spacer region[7], ERIC, while 16S rDNA[8] is the most popular one. Among the eubacterial 16S rDNA genes, the highly conserved sequences compose the constant regions, and the relatively less conserved sequences compose the variable regions, both interlace along the linear genes. Therefore, the pair of universal primers was carefully designed based upon the constant regions of 16S rDNA, so that they were capable of amplifying the 16S rDNA genes of all bacteria under certain circumstances. Meanwhile, the oligonucleotide probes were wisely designed based upon the variable regions of 16S rDNA at the species or genera level.
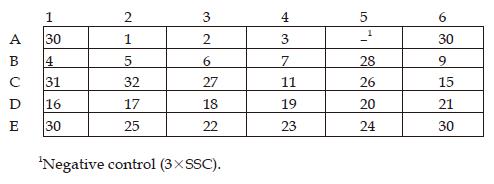
The standard strains used in this study including a wide range of species and many of the common organisms causing intestinal disease are listed in Table 1. These organisms were identified by conventional methods.
| Genus or species | Standard strain(s)1 |
| Salmonella | 50 001, 50 004, 50 009, 50 013, 50 014, 50 018, 50 019, 50 020, 50 021, 50 023, 50 029, 50 041, 50 042, 50 043, 50 047, 50 051, 50 073, 50 082, 50 083, 50 086, 50 093, 50 096, 50 098, 50 099, 50 100, 50 104, 50 105, 50 106, 50 109, 50 112, 50 115, 50 120, 50 124, 50 128, 50 145, 50 191, 50 200, 50 201, 50 220, 50 304, 50 306, 50 307, 50 309, 50 310, 50 313, 50 315, 50 320, 50 321, 50 322, 50 326, 50 327, 50 333, 50 335, 50 337, 50 338, 50 354, 50 355, 50 358, 50 360, 50 362, 50 402, 50 707, 50 708, 50 709, 50 710, 50 711, 50 712, 50 718, 50 719, 50 730, 50 731, 50 732, 50 733, 50 735, 50 736, 50 739, 50 746, 50 761, 50 774, 50 783, 50 825, 50 835, 50 846, 50 853, 50 854, 50 864, 50 913 |
| Shigella | 51 081, 51 100, 51 207, 51 227, 51 233, 51 252, 51 253, 51 255, 51 258, 51 259, 51 262, 51 307, 51 315, 51 334, 51 335, 51 336, 51 424, 51 464, 51 570, 51 571, 51 572, 51 573, 51 575, 51 582, 51 583, 51 584, 51 585, 51 610 |
| Escherichia coli | 44 102, 44 105, 44 109, 44 110, 44 113, 44 126, 44 127, 44 149, 44 155, 44 156, 44 186, 44 216, 44 336, 44 338, 44 344, 44 505, 44 710, 44 719, 44 752, 44 813, 44 824, 44 825 |
| Proteus | 49 027, 49 101, 49 102, 49 103 |
| Staphylococcus | 26 001, 26 003, 26 005, 26 101, 26 111, 26 113, 26 517 |
| Yersinia enterocolitica | 52 202, 52 203, 52 206, 52 207, 52 211, 52 215, 52 217, 52 219, 52 302 |
| Listeria monocytogenes | 54 003, 54 005, 54 006, 54 007 |
| Vibrio | 20 502, 20 506, 20 507, 20 511, 02-12 |
| Enterococcus faecalis | 32 221, 32 223 |
| Campylobacter jejuni | 26 277 |
| Bacillus cereus | 63 301 |
One colony from a fresh culture was resuspended in 100 μL distilled water in Eppendorf tubes. Then the tubes were transferred to a thermal cycler (Techgene, Techne Ltd.) and heated to 95 °C for 10 min. Finally, they were spun at 10 000 g for 1 min in a microcentrifuge, and 2 μL of the supernatant was used in PCR described below.
Strains divided from Hai River, Luan River, municipal sewage, and food samples from markets were used in this study. All the divided strains were identified by conventional methods and the VITEK test system (BioMerieux SA, France). Extraction of DNA from divided bacterial cultures was performed as above.
We downloaded 113 bacterial 16S rDNA sequences from the GenBank database. Then, we used the program ClustalW alignment of the software MacVector 6.5.1 to analyze these sequences and showed the conserved regions of 16S rDNA. The primers were based on the conserved regions 8 and 10 of the 16S rDNA. The sequences of forward primer 1169U20 (5’-AACTGGAGGAAGGTGGGGAT) and reverse primer 1521L19 (5’-AGGAGGTGATCCAACCGCA) were used to amply bacterial 16S rDNA. The forward primer 1169U20 was labeled with 5’-Cy3 fluorescence.
Sequences of forward primer invA-139 (5’-GTGAAATTATCGCCACGTTCGGGCAA) and reverse primer invA-141 (5’-TCATCGCACCGTCAAAGGAACC) were used to amplify the invA gene of Salmonella. Sequences of forward primer virA-1 (5’-CTGCATTCTGGCAATCTCTTCACATC) and reverse primer virA-2 (5’-TGATGAGCTAACTTCGTAAGCCCTCC) were used to amplify the virA gene of Shigella. The forward primers invA-139 and virA-1 were labeled with 5’-Cy3 fluorescence.
Each 50 μL reaction contained 33 μL sterile water, 5 μL 10×buffer (Takara Biotechnology Co., Ltd.), 2 μL supernatant from the extraction of bacterial DNA, 200 μmol/L dNTP mixture (Takara), 0.02 U/μL Takara Taq (Takara, 5 U/mL) and 0.1 μmol/L each primer (1169U20, 1521L19, invA-139, invA-141, virA-1, and virA-2). The PCR mixtures were subjected to 95 °C for 5 min, followed by 35 cycles at 94 °C for 25 s, at 55 °C for 30 s, and at 72 °C for 25 s. The PCR products were checked using 2% agarose electrophoresis and visualized with ethidium bromide staining.
All the oligonucleotide probes were chosen based on the variable regions between PCR primers using the alignment information. They were synthesized and modified with 3’-NH2 in order to increase their binding to the glass slide surface and their hybridization intensity.
Before use, the glass slides for microscopy must be cleaned as described by Brown (http://cmgm.stanford.edu/pbrown/protocols.html). Then the oligonucleotide probes were bound to the slides as follows: 5 μL of 50 μmol/L oligonucleotide drop was spotted on the glass slide by an arrayer (PixSys 5500 Workstation, Cartesian Technologies), 5 mm between each oligonucleotide spot. When all the oligonucleotide probes were applied, the glass slides were left at room temperature for 24 h to permit thorough drying of the DNA on the slide surface. After drying, the slides were washed twice in 0.2% SDS for 5 min each and twice in distilled water for 5 min each. Subsequently, the slides were washed in sodium borohydride solution (1.3 g Na2BH4 was dissolved in 375 mL phosphate buffered saline, and then 125 mL pure ethanol was added) for 5 min, in 0.2% SDS for 2 min, and twice in distilled water for 2 min each.
The fluorescent-labeled amplicons were hybridized to the oligonucleotide microarrays using the following protocol: 1 μL of amplicons was added into a tube containing 4 μL hybridization solution (UniHybTM, TeleChem International, Inc.), and the tube was heated to 95 °C for 10 min and was put on ice immediately. Mixture in the tube was then transferred onto the microarray, kept at 50 °C for 1 h in a hybridization cassette (TeleChem International, Inc.). After hybridization, unbound fluorescent amplicons were washed with washing buffer A (1×SSC+0.2% SDS) for 1 min, B (0.1×SSC+0.2% SDS) for 1 min, C (0.1×SSC) for 1 min, respectively.
We used the ScanArray 3000 (GSI Lumonics) to scan the area of the slide containing the microarray. The laser power and PMT were set at 80%.
The resolution of the ScanArray 3000 scanner is 10 μm, and the fluorescent density of each pixel is saved into the TIFF image file, facilitating further process and analysis of software. We uploaded the scanned image TIFF file into the ImaGene 4.0 software (BioDiscovery) to examine each feature for fluorescence intensity.
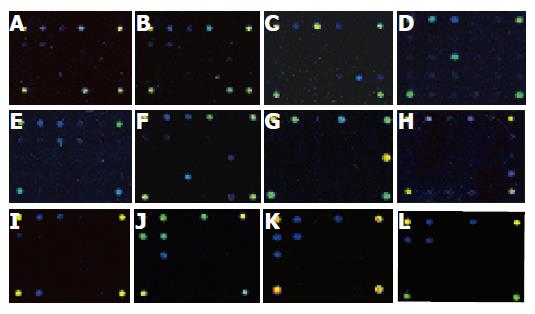
Under the same conditions for PCR amplification and hybridization, all the 170 strains produced PCR products, showing bands at approximately 370 bp, being equivalent to the fragment of 16s rDNA. Besides, strains belonging to Salmonella sp. produced another band of 285 bp, and strains belonging to Shigella sp. produced a 215-bp band. After the hybridization between PCR products and oligonucleotide probes, respective hybridization maps were built through the signal acquisition step using the ScanArray 3000 scanner. The original images generated by ScanArray 3000 scanner are shown in Figure 1. Monochrome fluorescent signals were mapped into pseudocolor spectrum according to their density in ascending order, e.g. black, dark blue, blue, green, yellow, red, white. Twelve typical hybridization maps corresponded to nine genera or species of bacteria, specifically.
The wisely designed oligonucleotide probes could be classified by their efficacy into six categories (Table 2). Category one, including oligonucleotide probe 1, a universal probe targeting the portion of 16S rDNA shared by all known eubacteria, was used to detect all kinds of known eubacteria. Category two, including oligonucleotide probes 2 and 3, the specific probes targeting the portion of 16S rDNA shared by both Gram-positive (G+) and negative (G–) bacteria, was used to distinguish between G+ and G– bacteria. Category three, including oligonucleotide probes 4 and 5, targeting the portion of 16S rDNA shared by all enteric bacteria, was used to identify intestinal bacteria at the family level. Category four, including oligonucleotide probes 6, 7, 9, 11, and 15-28, a cluster of genus or species-specific probes targeting the portion of 16S rDNA shared by their respective bacteria, was used to identify bacteria at genus or species level. Category five, including oligonucleotide probes 31 and 32, targeting the portion of the specific pathogenic genes of Shigella and Salmonella, was used to discriminate between bacteria of these two genera. Category six, including oligonucleotide probe 30, a positive control probe, was used both as a gauge to reflect the effectiveness of this hybridization system and as a reference coordinate for scanning. From hybridization signals of the five categories of oligonucleotide probes, it was easy to identify the target pathogenic bacteria in a given specimen. For instance, strong hybridization signals at the sites corresponding to the oligonucleotide probes 1, 3, and 23 were found, so the pathogenic bacteria in the given specimen could be sequentially identified as eubacteria, G– bacteria, and strains of Vibrio parahaemolyticus (Figure 1G). Some slightly weaker signals due to unspecific hybridization were also found. By means of multiple experiments, a hybridization signal was regarded as a specific signal if it meets the following criterion: the foreground fluorescent signal at an oligonucleotide probe spot was stronger than its background fluorescent signal with a signal-noise ratio larger than 100 calculated by ImaGene 4.0. Since the fluorescent signals of specific hybridization were three-fold stronger compared to those of unspecific hybridization, it was easy to identify the specific hybridization signals from the hybridization maps directly (Figure 2).
| No. | Sequence (5' to 3') | Target |
| 1 | gtacaaggcccgggaacgtattcacc | All known eubacteria (universal bacterial probe) |
| 2 | gacataaggggcatgatgatttgacgt | All Gram-positive bacteria |
| 3 | gtcgtaagggccatgatgacttgacgt | All Gram-negative bacteria |
| 4 | gtcatgaatcacaaagtggtaagcgc | All enteric bacterial |
| 5 | acgacgcactttatgaggtccgcttg | Escherichia coli, Shigella sp. and Salmonella sp. |
| 6 | gctcctaaaaggttactccaccggct | Staphylococcus aureus |
| 7 | cgacggctagctccaaatggttactg | Coagulase-negative Staphylococcus |
| 9 | tcacggtcttgcgtcttattgtacctac | Clostridium botulinum |
| 11 | gaactgagactggtttttaagtttggct | Clostridium perfringens |
| 15 | cgaactgggacatattttatagatttgc | Campylobacter jejuni |
| 16 | aggtcgccccttcgccgccctctgtatc | Legionella pneumophila |
| 17 | cgatccgaactgagaccggcttttaagg | Mycobacterium tuberculosis |
| 18 | tactcgtaagggccatgatacgacttaa | Proteus sp. |
| 19 | cgcggcttggcaaccctttgtaccgacc | Pseudomonas aeruginosa |
| 20 | actgagaatagttttatgggattagg | Listeria monocytogenes |
| 21 | gctccaccttcgcggtattcgctgccct | Vibrio cholerae |
| 22 | tcactttcgcaagttggccgccctctgt | Vibrio fluvialis |
| 23 | tggtaagcgtccccccgtagttgaaac | Vibrio parahaemolyticus |
| 24 | tacgacagactttatgtggtccgcttgc | Yersinia enterocolitica |
| 25 | cctcgcggtctagcagctcgttgtgctt | Enterococcus faecalis |
| 26 | ggattcgctcactatcgctagcttgcag | Aeromonas hydrophila |
| 27 | ccgacttcgggtgttacaaactctcg | Bacillus cereus, P. |
| 28 | gcttcatgcactcgagttgcagagtg | cocovenenans subsp. farinofermentans |
| 30 | atccccaccttcctccagtt | Positive control |
| 31 | cccccagaggcagagattgca | virA gene of Shigella sp. |
| 32 | cgccaataacgaattgcccga | invA gene of Salmonella sp. |
Twenty-six unselected divided cultures were also processed and hybridized as described above, and then identified according to the specific hybridization maps (Table 3). Among them, 25 strains were distinguished according to their hybridization maps, but only one strain was indistinguishable due to its weak signal. The comprehensive identification results by classical methods were regarded as the final standards. Except for sample 7, all other results were consistent with those detected by hybridization assay and conventional method, the consistency was 96.2% (25/26).
| No. | Hybridization assay | Conventional methods | Consistency |
| 1 | Staphylococcus aureus | Staphylococcus aureus | Y1 |
| 2 | Coagulase-negative Staphylococcus | Staphylococcus epidermidis | Y |
| 3 | Staphylococcus aureus | Staphylococcus aureus | Y |
| 4 | Staphylococcus aureus | Staphylococcus aureus | Y |
| 5 | Staphylococcus aureus | Staphylococcus aureus | Y |
| 6 | Pseudomonas aeruginosa | Pseudomonas aeruginosa | Y |
| 7 | -3 | Salmonella typhimurium | -2 |
| 8 | Escherichia coli | Escherichia coli | Y |
| 9 | Staphylococcus aureus | Staphylococcus aureus | Y |
| 10 | Pseudomonas aeruginosa | Pseudomonas aeruginosa | Y |
| 11 | Shigella sp. | Shigella flexneri | Y |
| 12 | Shigella sp. | Shigella flexneri | Y |
| 13 | Shigella sp. | Shigella flexneri | Y |
| 14 | Shigella sp. | Shigella flexneri | Y |
| 15 | Vibrio parahaemolyticus | Vibrio parahaemolyticus | Y |
| 16 | Yersinia enterocolitica | Yersinia enterocolitica | Y |
| 17 | Pseudomonas aeruginosa | Pseudomonas aeruginosa | Y |
| 18 | Shigella sp. | Shigella flexneri | Y |
| 19 | Shigella sp. | Shigella flexneri | Y |
| 20 | Vibrio parahaemolyticus | Vibrio parahaemolyticus | Y |
| 21 | Escherichia coli | Escherichia coli | Y |
| 22 | Escherichia coli | Escherichia coli | Y |
| 23 | Salmonella sp. | Salmonella typhimurium | Y |
| 24 | Campylobacter jejuni | Campylobacter jejuni | Y |
| 25 | Salmonella sp. | Salmonella typhimurium | Y |
| 26 | Salmonella sp. | Salmonella typhimurium | Y |
Our study showed that the gene chip-based method could identify a wide range of intestinal pathogenic bacterial species. The genus or species specific probes on the microarray are targeted at the 16S rDNA, while two discriminative probes are targeted at special pathogenic genes. Sample DNA was labeled with fluorescence by PCR, then hybridized to the probes on the chip, thus a couple of genus or species-specific hybridization patterns could be generated and used to discriminate the bacteria[9].
When the oligonucleotide probes for 16S rDNA were designed, we preferred a longer oligonucleotide segment with multiple mutation sites over a shorter one, which is also suitable for detecting single-nucleotide mutation[10-12]. The advantages are obvious: the amount of essential oligonucleotides is less, the cost of experiment is lower, and the identification of hybridization map is easier. However, the shortcoming is somewhat less discriminative to those sequences where only minor differences are present.
The 16S rDNA sequences of Shigella, Salmonella, and Escherichia coli are similar, and share almost identical sequences of the target 16S rDNA genes. Thus, these three genera can hardly be identified by only 16S rDNA[13,14].
Besides, intestinal pathogenic bacteria belonging to Listeria monocytogenes, Vibrio parahaemolyticus, Proteus sp., Vibrio cholerae, Enterococcus faecalis, Yersinia enterocolitica, and Campylobacter jejuni, could be detected and identified using the gene chip.
Compared to the classic microbial assay, immunological assay, PCR-based assay, the method based upon the gene chip technology could detect and identify a given strain of bacterium within 3 h. It is a fundamental start point to develop other methods for a large-scale assay. The target spectrum of this gene chip may be gradually expanded by adding newly designed oligonucleotide probes into the oligonucleotide microarray, and the accuracy may also be improved by increasing and readjusting the oligonucleotide probes in the oligonucleotide microarray. The arrangement of the oligonucleotide microarray may be rearranged according to its end usage. This method for intestinal pathogen assay using gene chip technology can be used for the diagnosis of infectious diseases, environmental supervision, food quality surveillance, etc.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999;21:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 359] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 2. | Bowtell DD. Options available--from start to finish--for obtaining expression data by microarray. Nat Genet. 1999;21:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 271] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 3. | Holloway AJ, van Laar RK, Tothill RW, Bowtell DD. Options available--from start to finish--for obtaining data from DNA microarrays II. Nat Genet. 2002;32 Suppl:481-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 140] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 4. | Chizhikov V, Rasooly A, Chumakov K, Levy DD. Microarray analysis of microbial virulence factors. Appl Environ Microbiol. 2001;67:3258-3263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 162] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 5. | Ludwig W, Schleifer KH. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 185] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 6. | Anthony RM, Brown TJ, French GL. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J Clin Microbiol. 2000;38:781-788. [PubMed] |
| 7. | Sharples GJ, Lloyd RG. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503-6508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 111] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Dams E, Hendriks L, Van de Peer Y, Neefs JM, Smits G, Vandenbempt I, De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16 Suppl:r87-173. [PubMed] |
| 9. | Call DR, Borucki MK, Loge FJ. Detection of bacterial pathogens in environmental samples using DNA microarrays. J Microbiol Methods. 2003;53:235-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 152] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 10. | Noller HF, Green R, Heilek G, Hoffarth V, Hüttenhofer A, Joseph S, Lee I, Lieberman K, Mankin A, Merryman C. Structure and function of ribosomal RNA. Biochem Cell Biol. 1995;73:997-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 36] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Boyer SL, Flechtner VR, Johansen JR. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol. 2001;18:1057-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 12. | Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat Genet. 1999;21:42-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 314] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 13. | Villalobo E, Torres A. PCR for detection of Shigella spp. in mayonnaise. Appl Environ Microbiol. 1998;64:1242-1245. [PubMed] |
| 14. | Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, Curtiss R, Gyles CL. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 651] [Article Influence: 19.7] [Reference Citation Analysis (0)] |