INTRODUCTION
Hepatitis C virus (HCV) infection is a major health care problem around the world[1]. It is estimated that there are about 170 million chronic carriers worldwide with four million in the USA[2-4] and five million in the western Europe[5-6]. HCV infection accounts for 20% of cases of acute hepatitis, 70% of chronic hepatitis, 40% of end-stage cirrhosis, 60% of cases of hepatocellular carcinoma, and 30% of liver transplantation[7-10]. Liver lesions are thought to be mainly related to immune-mediated mechanisms[5] but the exact mechanisms during HCV infection are still unclear[8,9,11]. However, the HCV core protein is a major component of viral nucleocapsid and it is a multifunctional protein that affects transcription and cell growth[12]. HCV core protein plays an important role in the HCV pathogenesis[13].
The ability of peroxisome proliferators to activate a receptor in the steroid receptor superfamily was first discovered in 1990[14], and the cognate protein was designated as peroxisome proliferator-activated receptors (PPARs). The PPARs are soluble transcription factors that are activated by a diverse class of lipophilic compounds[15]. With the activation of PPAR, a concomitant induction of a number of genes that code for peroxisomal fatty acid metabolizing enzymes was observed in mouse liver. There are three PPAR subtypes: PPARα (NR1C1), PPARβ (NR1C2), and PPARγ (NR1C3); and each subtype is capable of binding to DNA after heterodimerizing with RXR (NR2B1). PPARα is highly expressed in the liver, kidney, and cardiac smooth muscles. Many of the genes regulated by PPARα are involved in fatty acid metabolism[16]. PPARα could influence fatty acid import into hepatocyte mitochondria by up-regulating the expression of the liver-predominant mitochondrial carnitine palmtoylacyl-CoA transferase 1 (CPT1) gene[17].
Three CPT1 isoforms with various tissue distributions and encoded by distinct genes have been identified: CPT1A or L-CPT1 (liver isoform), CPT1B or M-CPT1 (muscle isoform), and CPT1C (brain isoform). CPT1A is expressed in the liver, the neonatal heart, and a number of other tissues, and has been the most investigated member of the acyltransferase family. It is anchored in the mitochondrial outer membrane by two transmembrane segments (TM1 and TM2), its N terminus (residues 1-47) and C-terminal catalytic domain (residues 123-773) being located on the cytosolic face of mitochondria. The N-terminal domain (1-147 residues) was shown to be essential for mitochondrial import and for the maintenance of a folded active and malonyl-CoA-sensitive conformation[18].
Until now, the role of PPARα and its target gene CPT1A in liver diseases is still limited to animal studies. In order to elucidate the mechanism of PPARα during the pathogenesis of HCV infection, PPARα and CPT1A expression levels were studied ex vivo and in vitro, respectively in this study.
MATERIALS AND METHODS
Subjects and liver samples processing
Chronic hepatitis C infection was defined by increased serum alanine transaminase activity (>35 IU/L), positive serum HCV replication determined by polymerase chain reaction (PCR; Amplicor Monitor v2.0; Roche, Indianapolis, IN, USA), and histological hepatic injury quantified by the METAVIR score evaluating the intensity of fibrosis (F1-F4) and necroinflammatory activity (A1-A3)[19]. The subjects were excluded if their body mass index was greater than 30 or if they suffered from diabetes or dyslipidemia.
Patient liver samples were collected from 46 patients with chronic HCV infection (41 transcutaneous needle liver biopsy samples from chronic hepatitis C patients and 5 surgical liver samples from HCV-related cirrhotic patients when they underwent transplantation). Non-HCV-infected liver tissues as controls were collected from 40 patients (29 normal tissue samples from patients who underwent surgery because of hepatic carcinoma, 5 biopsy samples from patients with alcoholic cirrhosis, and 6 samples from patients with alcoholic hepatitis). Both patients and controls did not receive any therapy (antiviral therapy, hepatotoxic drugs, corticosteroids, or immunosuppressive drugs) before or during liver sample collection.
The liver samples collected from hepatitis C patients or controls were divided into two parts: one part was fixed in 4% paraformaldehyde/phosphate-buffered saline and embedded in paraffin wax, and routinely processed for pathological analysis and for immunostaining[20]. Specimen slides were incubated for 48 h at 37 °C and then deparaffinized by dimethylbenzene and rehydrated by graded alcohol series, then stained with H&E, safran, and Masson’s trichrome. The other part was frozen immediately in the liquid nitrogen and stored at -80 °C for mRNA and protein analysis.
HepG2 cell culture and transfection
Human hepatocellular carcinoma cell line HepG2 was used for this study. Cells were maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (Eurobio, Les Ulis, France) in a humidified atmosphere of 50 mL/L CO2 in air at 37 °C.
In order to analyze the effect of the HCV core protein on PPARα expression, HepG2 cells were transfected with vector pEF352neo carrying, under the control of the elongation factor-1α promoter, an HCV complementary DNA including 1b HCV sequences from core to NS3 region as previously described[21]. Two independent clones (clone N3 and N4) stably expressing HCV core protein were analyzed. The clone transfected with the empty vector acted as negative control. Cells were resuspended in lysis buffer with 10% β-mercaptoethanol for RNA isolation[22]. All studies were performed in triplicate samples for three separate experiments.
RNA extraction and the real-time PCR analysis
Total RNA was extracted from cells and liver tissues by RNeasy kit (Macherey Nagel, Hoerdt, France) and TRIzol reagent (Life Technologies, Cergy Pontoise, France) following the protocols provided by the manufacturers with some modification[20]. RNA quantification was performed using spectrophotometry. After treatment at 37 °C for 30 min with 20-50 U of RNase-free DNase I (Roche Diagnostics Corporation), oligo-dT primers (Roche Diagnostics Corporation) were used to synthesize single-stranded complementary DNA. PPARα and CPT1A mRNAs were quantified using SYBR Green Master Mix (Applera, Courtaboeuf, France) with specific primers in a GeneAmp ABI prism 7000 (Applera). The primers used were as follows: PPARα anti-sense 5’-CCA CCA TCG CGA CCA GAT-3’, PPARα sense 5’-GAC GTG CTT CCT GCT TCA TAG A-3’; CPT1A anti-sense 5’-TGT GCT GGA TGG TGT CTG TCT C-3’, CPT1A sense 5’-CGT CTT TTG GGA TCC ACG ATT-3’; TBP anti-sense 5’-TTT TCT TGC TGC CAG TCT GGA C-3’, TBP sense 5’-CAC GAA CCA CGG CAC TGA TT-3’. Calibrated and nontemplate controls were included in each assay. Each sample was run in triplicate. SYBR Green dye intensity was analyzed using the ABI prism 7000 SDS software (Applera). All results were normalized to the TATA box-binding protein, an unaffected housekeeping gene[23]. All quantifications were performed in triplicate samples for three separate experiments.
Western blotting analysis
Protein preparation and immunoblotting were performed in liver specimens as described before. Total protein extracts were obtained by homogenization of tissues in an extraction buffer containing phosphate-buffered saline with 1% tergitol NP-40, 0.5% sodium desoxycholate, 0.1% sodium dodecyl sulfate, and a classic protease inhibitor cocktail. One hundred micrograms of total proteins were then separated by 10% polyacrylamide gel electrophoresis and electroblotted. Blots were blocked for 1 h at 4 °C with 5% milk Tris-Tween buffered saline 1×, and were incubated overnight at 1:1 000 with the anti-PPARα (Geneka Biotechnology Inc., Montreal, Canada). Membranes were incubated for 1 h with a swine anti-rabbit IgG conjugated to horseradish peroxidase for PPARα (dilution 1:1 000, Dako Laboratories, Trappes, France). Protein bands were revealed by an enhanced chemiluminescence reaction for PPARα. The results were expressed as units of optical density per quantity of total protein[20,24,25].
Immunohistochemical staining in liver tissues
Serial sections of paraffin embedded liver tissues were cut at 4 μm by Leica RM2145 rotary microtome (Leica Instruments GmbH, Germany). Glass slides were treated with poly-L-lysine in advance. Specimen slides were incubated for 48 h at 37 °C and then deparaffinized by xylene and rehydrated by graded alcohol series. An immunohistochemical staining was performed[20,24,25] for the detection of PPARγ expression. Liver sections were incubated in 3% H2O2 in methanol to deactivate endogenous peroxidase activity for 20 min. Then slides were incubated with 0.05% Saponin (ICN Biomedicals, OH, USA) for 30 min at room temperature to permeablize the tissues. After being washed, sections were pre-treated with Avidin D, followed by Biotin (Vector Laboratories) to block non-specific binding of Biotin/Avidin system reagents. Non-specific antibody binding was blocked with 1.5% goat serum in PBS for 15 min. Slides were incubated with 5% milk and 0.1% bovine serum albumin (BSA) in PBS for 15 min. Sections were incubated with the primary goat polyclonal antibody directed against PPARγ at room temperature (dilution 1:50, TEBU International) for 30 min, and were processed for peroxidase immunostaining using the Dako Laboratories system following the manufacturer’s instructions. Sections were incubated for 30 min at room temperature in rabbit anti-goat IgG (dilution 1:100, Dako Laboratories), and then under the same conditions in an avidin-biotinylated peroxidase complex (Dako, Denmark) that was prepared at least 30 min before use. Staining was developed with 3,3’-diaminobenzidine in chromogen solution for 1 min, and the reaction was stopped in distilled water. Sections were counterstained with hematoxylin. As a negative control experiment, the primary antibody was omitted and replaced with a non-specific antibody[20].
Statistical analysis
All results were expressed as mean±SD. Comparisons were analyzed by the nonparametric Mann-Whitney U test. Differences were considered statistically significant, if the P value was less than 0.05.
RESULTS
Patients’ ALT, HCV load, histological scores, and genotype
In this study, the body weights of the chronic hepatitis C patients were within the normal range and homogeneous. Before and during the study, they did not receive any treatment which could have influenced the results. Their mean serum alanine transaminase value was 119±27 IU/L, mean peripheral blood HCV RNA level was 747±267×103 IU/mL, and their histological lesions were consistent at the time of liver biopsy examination. Half of the patients had fibrosis scored F1, 24% F2, and 26% F3-F4. Seventy-two percent of the cases had an inflammatory activity scored A1 and 28% A2. Fifty-two percent of patients were genotype 1, 31% were genotype 3, 12% were genotype 2, and 5% were genotype 4.
HCV infection reduced the expression of PPARα and CPT1A
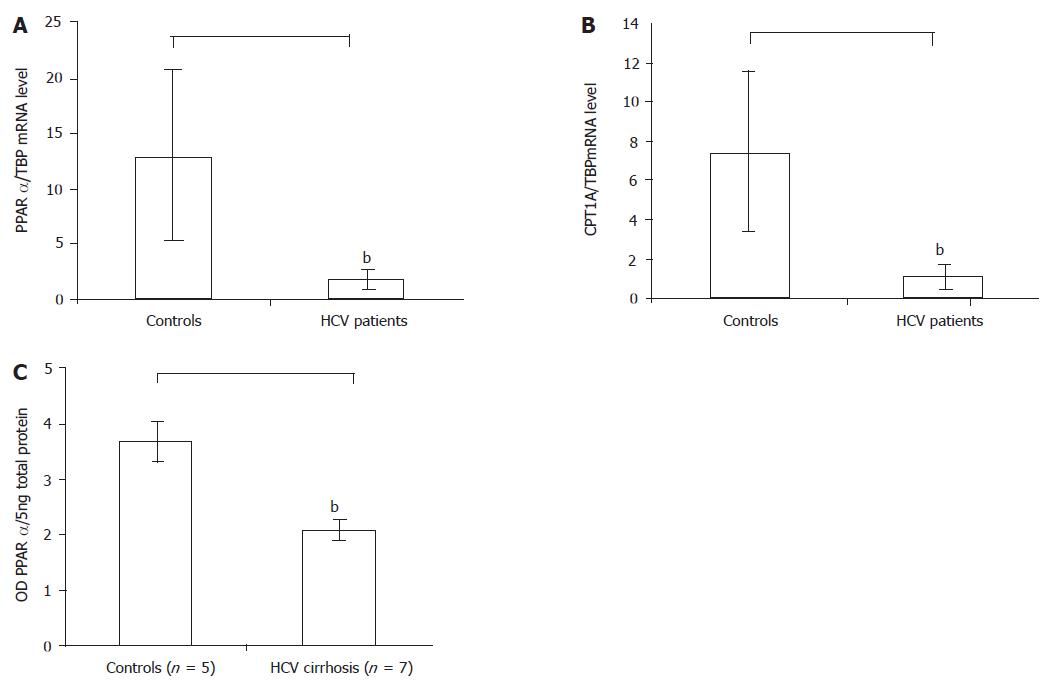
The real-time PCR analysis indicated that the housekeeping gene TATA box-binding protein concentrations were similar in patients and controls. But the level of PPARα in hepatitis C patient livers lowered by 86% compared with controls (1.8±2.8 vs 13±3.4, P = 0.0002) (Figure 1A). And the reduction in CPT1A mRNA was similar to the reduction in PPARα in the livers of patients in the study (Figure 1B). Compared with the 40 controls, the expression level of the CPT1A gene in the 46 hepatitis C patients lowered by more than 80% (1.1±1.5 vs 7.4±1, P = 0.004).
Figure 1 A: The PPARα mRNA level in the HCV patients reduced compared with controls by real-time RT-PCR, bP<0.
01 vs control; B: The PPARα target gene CPT1A mRNA level in the HCV patients also reduced compared with controls by real-time RT-PCR, bP<0.01 vs control; C: The PPARα protein level in the HCV cirrhotic patients reduced compared with controls by Western blot. Results were expressed as mean±SD, bP<0.01 vs control.
In a similar pattern, the Western blot also proved the results of real-time PCR analysis. Because of the limited amount of the liver biopsy samples, the Western blot analysis of PPARα was performed only in the surgically obtained liver specimens of patients with HCV infection (n = 5) and controls (n = 7). The analysis revealed a band with a molecular weight of approximately 55 ku corresponding to PPARα in all surgical liver specimens, and significantly lower levels of PPARα protein in HCV patients than in controls (2.3±0.3 vs 3.6±0.2 OD of PPARα protein/5 ng total protein, P = 0.009) (Figure 1C).
PPARα protein expression decreased in hepatocytes
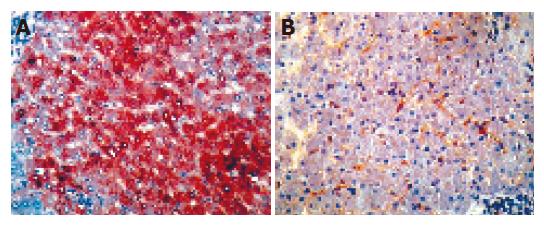
The immunohistochemical staining was performed in all hepatitis C patients’ and controls. Cell staining was positive for PPARα mainly in hepatocytes and by perinuclear staining (Figure 2, left). However, the results of stained cells in chronic hepatitis C patients indicated a decrease in PPARα staining in hepatocytes compared to those in control livers (Figure 2, right). Negative control staining was performed by omitting the primary antibody or the use of an irrelevant antibody. These results suggested that hepatocytes were the major sources of PPARα and they expressed low levels of PPARγ during HCV infection.
Figure 2 Representative PPARα immunostainings in liver specimens.
The left: a control liver, PPARα staining was detected in the majority of hepatocytes (magnification ×160); The right: a severe hepatitis patient liver, the number of PPARα-stained hepatocytes was decreased markedly (magnification ×160).
HCV core protein reduced the expression of PPARα
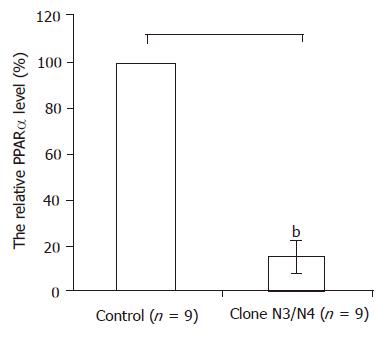
Because hepatocytes were the parenchymal component in the liver that were prone to be infected by HCV, and HCV core protein regulated the transcriptional activity of several genes, the PPARα expression in HepG2 cells stably expressing HCV core protein were also quantified with the real-time PCR. In the N3 and N4 colons which were stably expressing the HCV core protein, PPARα mRNA levels were found to be similar, but decreased more than 80% compared with controls (Figure 3).
Figure 3 In the N3 colon and N4 colon which were stably expressing the HCV core protein PPARα mRNA levels decreased more than 80% compared with controls by real-time RT-PCR.
Results were expressed as mean±SD, bP<0.01 vs control.
DISCUSSION
Because the morbidity and mortality of hepatitis C is high and the natural history is generally long and slow, many patients develop complications[6-8]. It is difficult to find an effective treatment if the exact mechanism of HCV is not clear, so it is a very important task to elucidate the pathogenesis of hepatitis C[2,4]. The results in this study demonstrated for the first time that the expression of PPARα was impaired in the livers of chronic hepatitis C patients. Both in vitro studies and the transgenic mouse model suggested that the HCV core protein is possibly responsible for lipid accumulation[12,26]. It was reported that HCV core protein expression in transgenic mice could inhibit microsomal triglyceride transfer protein activity and very low density lipoprotein secretion causing steatosis[13]; and the mechanism postulated seemed to be related to mitochondrial toxicity with the production of reactive oxygen species. In this study, it was also found that in the N3 colon and N4 colon stably expressing the HCV core protein, the PPARα mRNA levels lowered significantly compared with controls. These results indicated the involvement of the HCV protein in the regulation of PPARα expression and activation.
PPARα plays a fundamental role in regulating energy homeostasis through controlling lipid metabolism[14]. PPARs belong to the type II steroid receptor family, including members such as RXR and thyroid hormone receptor[27]. These receptors are generally considered to localize to the nucleus and appear not to be bound to other proteins in inactive complexes, as has been extensively described for type I receptors (e.g. glucocorticoid receptor, progesterone receptor, androgen receptor). PPARα is a fatty acid-activated transcription factor that up-regulates the expression of a variety of genes encoding proteins involved in β-oxidation and lipoprotein metabolism[16]. PPARα plays a central role in fatty acid homeostasis by regulating the degradation of fatty acids by mitochondrial as well as by peroxisomal and microsomal fatty acid oxidation[28]. In addition, PPARα contributes to the maintenance of energy balance by regulating the expression of enzymes that participate in mitochondrial fatty acid oxidation and the formation of ketone bodies from fatty acids. Lack of this transcriptional factor in PPARα–/– mice results in the inability to up-regulate hepatic fatty acid oxidation and ketogenesis in the face of increased concentrations of free fatty acids in the circulation[15]. In this study, both the real-time PCR and Western blot analysis indicated that PPARα expression level was down-regulated in HCV infection; and the immunohistochemical staining also showed that the decreased expression was mainly in hepatocytes. So it suggests that HCV injures the liver partly through down-regulation of PPARα.
The PPARα-responsive human genes include CPT1A, the long chain fatty acyl-CoA synthase (ACS), and the mitochondrial HMG-CoA synthase (HMGCS2). Several aspects of fatty acid oxidation are disrupted in PPARα null mice that include a diminished, constitutive expression of several components of the mitochondrial fatty acid oxidation pathway. These results suggest that in mice, PPARα is a significant regulator of the mitochondrial capacity for fatty acid β-oxidation. The flux of long chain fatty acids through the mitochondrial γ-oxidation pathway is regulated by CPT1, a component of the long chain fatty acylcarnitine translocases. The activity of CPT1 is modulated by the concentration of malonyl-CoA, a potent inhibitor of the enzyme formed in the first committed step of fatty acid synthesis. Because CPT1 is induced by peroxisome proliferators in the liver, the target gene CPT1A of PPARα was also studied in order to understand the possible mechanisms of reduced expression of PPARα in the HCV infection. The human CPT1A gene is PPARα-responsive[17,18,29-31]. The fatty acyl-CoA substrates for CPT1 are formed by the esterification of free fatty acids to CoA by ACS. ACS is induced by peroxisome proliferators in rat liver, and PPRE has been identified in the promoter of the rat gene. In this study, the expression levels of the CPT1A gene in the livers of hepatitis C patients decreased significantly. It indicated that HCV core protein was possibly responsible for lipid accumulation by CPT1A, which was regulated by the transcriptional factor PPARα that was also the major isoform required for mediating the responses resulting from the actions of peroxisome proliferators in liver.
It was reported that human genes encoding key enzymes for mitochondrial fatty acid oxidation and ketogenesis could be regulated by human PPARα. In these tissues malonyl-CoA may act as a sensor or hormone and fuel through its ability to inhibit mitochondrial outer membrane CPT I[17]. This enzyme catalyzes the transesterification of long-chain fatty acyl-CoAs to long-chain acylcarnitines, which are carried into the mitochondrial matrix where acyl-CoA is regenerated for β-oxidation[18,30,31]. The role of PPARα in cell proliferation[32,33], tumor promotion[34,35] and in the prevention/reversal of liver injury has been studied by different researchers before[34-36].
To sum up, this report showed that the reduced intrahepatic PPARα expression level was associated with the HCV core protein. HCV infection down-regulated the expression of PPARα and CPT1A not only at mRNA level but also at the protein level. Correspondingly, the reduced PPARα level induced the low expression of CPT1A. The immunohistochemical analysis also proved that expression level of PPARα decreased in the hepatocytes. The function impairment of PPARα and CPT1A during the HCV infection still needs further investigation. The results in this study indicated that PPARα played an important role in the pathogenesis of chronic HCV infection, and this nuclear receptor could be a potential therapeutic target[34-36] in the treatment of HCV infection in the future.