Published online Nov 21, 2005. doi: 10.3748/wjg.v11.i43.6880
Revised: April 28, 2005
Accepted: April 30, 2005
Published online: November 21, 2005
AIM: To screen the immunogenic membrane proteins of Shigella flexneri 2a 2457T.
METHODS: The routine two-dimensional polyacrylamide gel electrophoresis (2-DE) and Western blotting were combined to screen immunogenic proteins of S. flexneri 2a 2457T. Serum was gained from rabbits immunized with the same bacteria. Immunogenic spots were cut out from the polyacrylamide gel and digested by trypsin in-gel. Matrix-assisted laser desorption/ionization time of flight-mass spectrometry (MALDI-TOF-MS) was performed to determine the molecular weight of peptides. Electrospray ionization (ESI-MS/MS) was performed to determine the sequences of the interesting peptides.
RESULTS: A total of 20 spots were successfully identified from Coomassie brilliant blue stained gels representing 13 protein entries, 5 known antigens and 8 novel antigens. A hypothetical protein (YaeT) was detected, which might be a candidate target of vaccine.
CONCLUSION: Membrane proteins of S. flexneri 2a 2457T were successfully observed by 2-DE. Several known and novel antigens were identified by mass spectrum.
- Citation: Ying TY, Wang JJ, Wang HL, Feng EL, Wei KH, Huang LY, Huang PT, Huang CF. Immunoproteomics of membrane proteins of Shigella flexneri 2a 2457T. World J Gastroenterol 2005; 11(43): 6880-6883
- URL: https://www.wjgnet.com/1007-9327/full/v11/i43/6880.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i43.6880
The genus Shigella spp. is a group of Gram-negative enteric bacilli which cause bacillary dysentery in human beings, accounting for 20% of the 4.6 million diarrhea-associated deaths among children[1]. Though the LPS can induce a good immune response in human beings, the role of proteins (especially the membrane proteins) in conferring immunity to shigellosis is at best speculative. Considering outer membrane proteins of Shigella spp. function as a dynamic interface between the cell and its surroundings, it is possible to develop new antigens from them. Due to the methodology limitations of protein separation and identification, it is difficult to identify the immunogenic proteins in bands on 1-D gel. With the improvement of 2-DE in recent years much valuable information is available and immunoproteomics has been built around 2-DE and routine immunologic technologies.
S. flexneri 2a is the dominant serotype causing shigellosis in China. Our laboratory has finished a two-dimensional electrophoresis reference map and a proteomic database of S. flexneri 2a 2457T[2], but only a few of membrane proteins can be identified in that database. In order to develop new protective antigens against S. flexneri and to understand their immune mechanism, we applied immunoproteomic technologies in screening new antigens of S. flexneri 2a 2457T.
S. flexneri 2a 2457T was aerobically cultured in LB overnight at 37 °C. Overnight cultures were diluted 1:100 and shaken at 250 r/min. Growth was stopped at the early stationary phase at an A600 of 3.3.
Cells were harvested and centrifuged for 15 min at 2 000 r/min (Sigma 3K12, No. 12150; St. Louis, MO, USA) at 4 °C. The pellet was washed thrice for 10 min at 2 000 r/min with low-salt washing buffer (3 mmol/L KCl, 1.5 mmol/L KH2PO4, 68 mmol/L NaCl, 9 mmol/L NaH2PO4)[3]. Proteins were extracted using the ReadyPrepTM protein extraction kit (Membrane I) (BioRad, USA). Integral membrane proteins were separated from hydrophilic proteins using the nonionic detergent Triton X-114.
Eighteen-centimeter immobilized pH gradient (IPG) strips (pH ranges, 4-7) (Amersham Pharmacia Biotech, Sweden) were used. Isoelectric focusing (IEF) was conducted for 60 000 Vh (IPGphor, Amersham Pharmacia Biotech). Vertical slab SDS-PAGE (12.5%) was run at 30 mA/gel for the second dimension. Gels were stained with Colloidal Coomassie Blue[4]. Image analysis was performed with Image-Master 2D Elite Version 3.1.
S. flexneri 2a 2457T was aerobically cultured in LB overnight at 37 °C. Rabbits were immunized six times with culture solution intravenously at intervals of 5 d. The doses were (5, 7.5, 10, 15, 20, 20)×108 CFU, respectively. Eight days after the last immunization, blood was collected from the tested animals and the sera were separated. Antibody titers 1:5 120 was measured by microaggalutination test and ELISA.
After two-dimensional electrophoresis, the gels were electroblotted onto Hybond™ECL™ nitrocellulose membrane (Amersham Pharmacia Biotech) using a semi-dry transfer unit (Hoefer™ TE 77, Amersham Pharmacia Biotech, Sweden). Before immunodetection, the membranes were stained for 10 min with 5 g/L Ponceau S in 10 mL/L acetic acid and the positions of some selected spots were marked by clean needles. Western blotting was performed as previously described[5]. Then antigen-antibody complexes were detected with peroxidase-labeled goat anti-rabbit IgGs and substrate.
In-gel protein digestion was performed as previously described[6]. All MALDI-MS measurements were performed on a Bruker Reflex. III MALDI-TOF-MS (Bruker Daltonik, Bremen, Germany) operating in reflectron mode.
The peptide solution after in-gel protein digestion was desalted with ZipTip C18TM pipette tips (Millipore, Bedford, MA, USA). Electrospray ionization (ESI-MS/MS) was carried out with a hybrid quadrupole orthogonal acceleration tandem mass spectrometer (Q-TOF2) (Micromass, Manchester, UK)[2].
Peptide mass fingerprinting searches were performed using the program Mascot developed by Matrix Science Ltd (http://www.matrixscience.com). For protein identification, peptide mass searches against the database of 2457T by Mascot licensed in-house and the searches against the NCBInr database with free access on the internet were done. A peptide mass accuracy of 0.3 Da was defined.
The sample was prepared on the basis of the separation of membrane proteins by temperature-dependent phase partitioning using Triton X-114 detergent. Proteins anchored to the membrane or containing one or two transmembrane domains were efficiently partitioned to the detergent-rich phase. In order to solubilize the protein thoroughly, thiourea was used. In pH 4-7 gradient 2-DE map, 148 spots were cut and 111 spots were successfully identified by MALDI-TOF-MS presenting 82 protein entries. Twenty-five proteins were not observed/identified in our previous work[2]. The majority of these 25 proteins (data not shown) were hydrophobic and associated with the membrane. The relative abundance of membrane-associated proteins identified in this study was higher than that in our previous study[2].
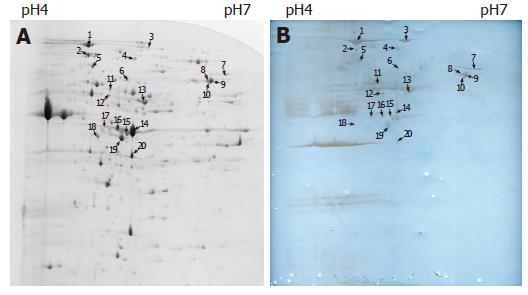
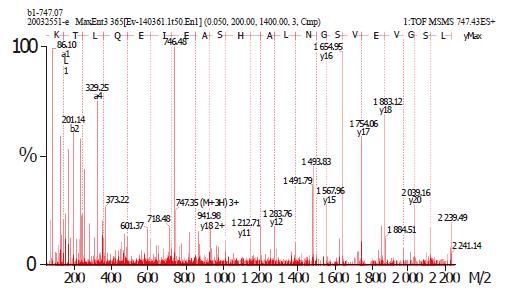
On the basis of the established immunoproteomic map of soluble proteins of S. flexneri 2a 2457T (unpublished), we described a group of spots in a 2-DE map of immunogenic proteins from hydrophobic proteins in this study. Five hundred micrograms of protein sample was used to perform the 2-DE. One of the parallel gels was electroblotted onto nitrocellulose membrane and the other was stained with Coomassie brilliant blue G-250. We successfully identified 20 immunoreactive spots from Coomassie brilliant blue stained gels using sera from immunized rabbits, which represented 13 protein entries, 5 known antigens and 8 novel antigens. The 20 spots were marked on the 2-D gel and corresponding blotting membrane (Figure 1). Table 1 lists all the identified proteins. ESI-MS/MS was used to confirm the protein marked as spot 1. Figure 2 shows the result of ESI-MS/MS identification.
| Spot ID | Gene symbol | Protein common name | NCBI GI identifier | Cellular role |
| 1 | YaeT | Hypothetical protein | gi|30061734 | Cell envelope |
| 2 | DnaK | Chaperone Hsp70; autoregulated heat shock protein | gi|30061584 | Protein fate |
| 3 | ClpB | Heat shock protein | gi|30063993 | Protein fate |
| 4/14/16/17/ 20 | OmpA | Outer membrane protein 3a (II*; G; d) | gi|30062494 | Cell envelope |
| 5 | MopA | GroEL, chaperone Hsp60, peptide-dependent ATPase, heat shock protein | gi|30065518 | Protein fate |
| 6 | Pgm | Phosphoglucomutase | gi|30062137 | Energy metabolism |
| 7 | OppA | Periplasmic oligopeptide binding protein | gi|30062764 | Protein fate |
| 8/9 | AtpA | Membrane-bound ATP synthase, F1 sector, alpha-subunit | gi|30064961 | Energy metabolism |
| 10 | LpdA | Lipoamide dehydrogenase (NADH) | gi|30061682 | Energy metabolism |
| 11 | Gnd | Gluconate-6-phosphate dehydrogenase | gi|30063478 | Energy metabolism |
| 12/13/18 | TufB | Protein chain elongation factor EF-Tu | gi|30064737 | Protein synthesis |
| 15 | Tsf | Protein chain elongation factor EF-Ts | gi|30061727 | Protein synthesis |
| 19 | MglB | Galactose-binding transport protein; receptor for galactose taxis | gi|30063593 | Transport and binding proteins |
Our results are in accordance with other studies[7-11].The outer membrane protein 3a (II*; G; d) is a precursor of OmpA, a major and highly conserved outer membrane protein of Gram-negative bacteria. Due to its high copies per cell[12], multiple charged isoforms[13] and its strong immunogenecity, identification of OmpA was performed several times during the immunoproteomics analysis. All these proteins were observed in our other works (unpublished).
Besides the above confirmatory findings, the study detected several new immunoreactive proteins (AtpA, OppA, MglB, LpdA, ClpB, Gnd, Pgm, YaeT). AtpA, LpdA, Gnd, and Pgm are components of the energy metabolism system. ATP synthesis/hydrolysis occurs in the ATP synthase F1 sector which lies at the surface of cytoplasmic membrane. LpdA codes for an outer membrane lipoamide dehydrogenase that is highly immunogenic. It is an in vivo-induced antigen in Mycobacterium tuberculosis[14]. Since LpdA is a functional subunit of both pyruvate dehydrogenase (aceEF) and alpha-ketoglutarate dehydrogenase (sucAB), a lpdA mutant of H. influenzae can be significantly attenuated[15]. Gnd is an important component of pentose phosphate pathway. Phosphoglucomutase (pgm) is associated with virulence of Brucella abortus because the deltapgm strain is unable to assemble the O side chain in the complete LPS. Vaccination with the deltapgm strain induces effective protection[16]. The periplasmic oligopeptide binding protein OppA is part of the oligopeptide transport system. In addition to the function mentioned above, it also plays a role in mediating the adhesion or interactions of bacteria to different substrates, tissues or environments[17-19]. OppA and periplasmic galactose-binding protein MglB also display some chaperone-like functions, suggesting that they are probably involved in protein folding and protection against stress in periplasm[20]. ClpB is also a heat shock protein. The proteins described above have not been reported as antigens and may serve as candidate markers for bacterial infection though they are unlikely to be protective.
A hypothetical protein (YaeT) detected is of high homology to Oma90 of S. flexneri M90T (serotype 5)[21]. We also detected this protein in another study (unpublished), which is verified by ESI-MS/MS. Since it has an enhanced expression in a murine model and exhibits strong homology to genes encoding Haemophilus influenzae D15 and Pasteurella multocida Oma87, its role in Shigella infection and immunoreaction is worthy to be clarified.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Ahmed F, Ansaruzzaman M, Haque E, Rao MR, Clemens JD. Epidemiology of postshigellosis persistent diarrhea in young children. Pediatr Infect Dis J. 2001;20:525-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Liao X, Ying T, Wang H, Wang J, Shi Z, Feng E, Wei K, Wang Y, Zhang X, Huang L. A two-dimensional proteome map of Shigella flexneri. Electrophoresis. 2003;24:2864-2882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Humphery-Smith I, Guyonnet F, Chastel C. Polypeptide cartography of Spiroplasma taiwanense. Electrophoresis. 1994;15:1212-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Cordwell SJ. Acquisition and archiving of information for bacterial proteomics: from sample preparation to database. Methods Enzymol. 2002;358:207-227. [PubMed] |
| 5. | Wu M, Stockley PG, Martin WJ. An improved western blotting technique effectively reduces background. Electrophoresis. 2002;23:2373-2376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Liao X, Ying TY, Wang HL, Wang J, Wei KH, Huang LY, Huang PT. A modified method of in-gel digestion of Coomassie brilliant blue-stained 2-D gels. Shengwu Jishu Tongxun. 2003;14:509-511. |
| 7. | Sanchez-Campillo M, Bini L, Comanducci M, Raggiaschi R, Marzocchi B, Pallini V, Ratti G. Identification of immunoreactive proteins of Chlamydia trachomatis by Western blot analysis of a two-dimensional electrophoresis map with patient sera. Electrophoresis. 1999;20:2269-2279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Bini L, Sanchez-Campillo M, Santucci A, Magi B, Marzocchi B, Comanducci M, Christiansen G, Birkelund S, Cevenini R, Vretou E. Mapping of Chlamydia trachomatis proteins by immobiline-polyacrylamide two-dimensional electrophoresis: spot identification by N-terminal sequencing and immunoblotting. Electrophoresis. 1996;17:185-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Pardo M, Ward M, Pitarch A, Sánchez M, Nombela C, Blackstock W, Gil C. Cross-species identification of novel Candida albicans immunogenic proteins by combination of two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Electrophoresis. 2000;21:2651-2659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | McAtee CP, Lim MY, Fung K, Velligan M, Fry K, Chow T, Berg DE. Identification of potential diagnostic and vaccine candidates of Helicobacter pylori by two-dimensional gel electrophoresis, sequence analysis, and serum profiling. Clin Diagn Lab Immunol. 1998;5:537-542. [PubMed] |
| 11. | Haas G, Karaali G, Ebermayer K, Metzger WG, Lamer S, Zimny-Arndt U, Diescher S, Goebel UB, Vogt K, Roznowski AB. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics. 2002;2:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 12. | Nikaido H. Outer membrane. Escherichia coli and Salmonella: Cellular and molecular biology. Washington DC: ASM Press 1996; 29-47. |
| 13. | Molloy MP, Herbert BR, Slade MB, Rabilloud T, Nouwens AS, Williams KL, Gooley AA. Proteomic analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871-2881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 363] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 14. | Deb DK, Dahiya P, Srivastava KK, Srivastava R, Srivastava BS. Selective identification of new therapeutic targets of Mycobacterium tuberculosis by IVIAT approach. Tuberculosis (Edinb). 2002;82:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 15. | Herbert M, Kraiss A, Hilpert AK, Schlör S, Reidl J. Aerobic growth deficient Haemophilus influenzae mutants are non-virulent: implications on metabolism. Int J Med Microbiol. 2003;293:145-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 16. | Ugalde JE, Comerci DJ, Leguizamón MS, Ugalde RA. Evaluation of Brucella abortus phosphoglucomutase (pgm) mutant as a new live rough-phenotype vaccine. Infect Immun. 2003;71:6264-6269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Sutcliffe IC, Russell RR. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123-1128. [PubMed] |
| 18. | Higgins CF, Hardie MM. Periplasmic protein associated with the oligopeptide permeases of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1983;155:1434-1438. [PubMed] |
| 19. | Fenno JC, Tamura M, Hannam PM, Wong GW, Chan RA, McBride BC. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect Immun. 2000;68:1884-1892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 76] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Richarme G, Caldas TD. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J Biol Chem. 1997;272:15607-15612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Robb CW, Orihuela CJ, Ekkelenkamp MB, Niesel DW. Identification and characterization of an in vivo regulated D15/Oma87 homologue in Shigella flexneri using differential display polymerase chain reaction. Gene. 2001;262:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |