Published online Oct 28, 2005. doi: 10.3748/wjg.v11.i40.6295
Revised: December 21, 2004
Accepted: December 23, 2004
Published online: October 28, 2005
AIM: To explore the propriety of providing hepatitis B virus (HBV) genotypes F and H with two distinct genotypes.
METHODS: Eleven HBV isolates of genotype F (HBV/F) were recovered from patients living in San Francisco, Japan, Panama, and Venezuela, and their full-length sequences were determined. Phylogenetic analysis was carried out among them along with HBV isolates previously reported.
RESULTS: Seven of them clustered with reported HBV/F isolates in the phylogenetic tree constructed on the entire genomic sequence. The remaining four flocked on another branch along with three HBV isolates formerly reported as genotype H. These seven HBV isolates, including the four in this study and the three reported, had a sequence divergence of 7.3-9.5% from the other HBV/F isolates, and differed by >13.7% from HBV isolates of the other six genotypes (A-E and G). Based on a marked genomic divergence, falling just short of >8% separating the seven genotypes, these seven HBV/F isolates were classified into F2 subtype and the former seven into F1 subtype provisionally. In a pairwise comparison of the S-gene sequences among the 7 HBV/F2 isolates and against 47 HBV/F1 isolates as well as 136 representing the other six genotypes (A-E and G), two clusters separated by distinct genetic distances emerged.
CONCLUSION: Based on these analyses, classifying HBV/F isolates into two subtypes (F1 and F2) would be more appropriate than providing them with two distinct genotypes (F and H).
- Citation: Kato H, Fujiwara K, Gish RG, Sakugawa H, Yoshizawa H, Sugauchi F, Orito E, Ueda R, Tanaka Y, Kato T, Miyakawa Y, Mizokami M. Classifying genotype F of hepatitis B virus into F1 and F2 subtypes. World J Gastroenterol 2005; 11(40): 6295-6304
- URL: https://www.wjgnet.com/1007-9327/full/v11/i40/6295.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i40.6295
Seven genotypes of hepatitis B virus (HBV) have been differentiated by a sequence divergence in the entire genome >8%, and they are named by capital alphabet letters from A to G[1-3]. Among the seven genotypes of HBV, F is the most divergent from the other six and separated from them by >14% in the full-length sequence. Recently, an eighth genotype was proposed for HBV isolates from Central America based on a sequence divergence from those of the seven genotypes (A-G) barely exceeding 8%[4].
An attempt was made in the present study to evaluate the independence of genotype H from the others by systematic phylogenetic analysis on the entire nucleotide sequences and their open reading frames of 25 HBV isolates of genotype F (HBV/F) or H, including the 11 from patients with acute or chronic hepatitis B from San Francisco, Japan, Panama, and Venezuela that were determined anew.
Because of a mediocre sequence divergence, a little short of clearing the >8% difference separating the seven HBV genotypes (A-G), taken along with their relationship in phylogenetical analysis, the two clades of HBV/F isolates would better be classified into two subtypes of genotype F, and they were provisionally assigned the designations F1 and F2. A possibility is proposed for distinguishing F1 and F2 subtypes by combining serotypes of the preS2-region product[5-7], for the determination of HBV genotypes, and subtypes of hepatitis B surface antigen (HBsAg).
Of the patients with acute or chronic hepatitis B who were taken care of in Gastroenterology Division of California Medical Center in San Francisco, Ryukyu University Hospital in Okinawa, Japan and National Hospitals in Panama and Venezuela, 11 were found to be infected with HBV/F. Demographic characteristics and liver diseases of the 11 patients are shown in Table 1. Their sera were tested for HBV markers, and HBV/F isolates in them were sequenced over the entire genome. The study design was approved by Ethics Committees of the institutions, and an informed consent was obtained from each patient.
| HBV strains | Accession number | Age (yr) | Gender | Ethnicity | Clinical diagnosis |
| USF10 | AB059659 | 46 | Female | Caucasian | Liver cirrhosis |
| USF1122 | AB064315 | 46 | Male | Hispanic | Liver cirrhosis |
| USF1778 | AB059660 | 44 | Male | Hispanic | Liver cirrhosis |
| USF2065 | AB059661 | 47 | Male | Hispanic | Acute hepatitis |
| USF2573 | AB064316 | 67 | Male | Hispanic | Liver cirrhosis |
| JPF1130 | AB086397 | 43 | Male | Asian | Chronic hepatitis |
| JPNTAKA | AB116654 | NA | Male | Asian | NA |
| VENEZ2 | AB116549 | 35 | Male | Hispanic | Acute hepatitis |
| VENEZ4 | AB116550 | 35 | Male | Hispanic | Acute hepatitis |
| PANAM5 | AB116551 | NA | NA | NA | NA |
| PANAM6 | AB116552 | NA | NA | NA | NA |
The seven major genotypes of HBV, i.e., A, B, C, D, E, F and G, were determined by ELISA with commercial kits (HBV Genotype EIA, Institute of Immunology, Tokyo, Japan) which distinguishes serotypes of the product of the preS2 region that are specific for each genotype[5-7]. It involves monoclonal antibodies directed to five preS2 epitopes designated b, k, m, s and u, respectively. Since the expression of preS2 epitopes are influenced by HBV genotypes, the combination thereof can define genotypes serologically; the epitope b is expressed regardless of HBV genotypes and guarantees the presence of preS2 product in the test serum. Thus, a serotype bsu is specific for genotype A, bm for B, bks for C and bk for F. Although genotypes D and E have the same serotype (bksu), they are differentiated by distinct behavior in the binding with another monoclonal antibody against the epitope g immobilized on a solid support. HBsAg of genotype E binds with immobilized anti-g, so that it is sandwiched between antibody to the common determinant a of HBsAg labeled with horseradish peroxidase, while HBsAg of genotype D is not. Although HBsAg of genotype G has a preS2 serotype for D, bksu(g), it has HBsAg subtype of adw unlike ayw in almost all HBsAg samples of genotype D. Thus, genotype G can be detected serologically by the combination of preS2 serotype bksu(g) and HBsAg subtype adw.
Nine sets of primers used in this study are shown in Table 2. The genomes of 11 HBV/F isolates were sequenced through 3 overlapping fragments; they were amplified by PCR with 3 sets of primers (HBV/Ff1-HBV/Fr1, HBV/Ff2-HBV/Fr2 and HBV/Ff3-HBV/Fr3). AmpliTaq Gold was activated at 96 °C for 9 min, and PCR was performed for 40 cycles (96 °C for 1 min; 55 °C for 1 min; 72 °C for 1.5 min [5 min in the last cycle]). The three nucleotide fragments were sequenced directly by the dideoxy method with use of the BigDye Terminator cycle sequencing kit in a fluorescent 3100 DNA sequencer (Applied Biosystems, Foster City, CA, USA) using the remaining six sets of primers along with the three primer sets used for PCR (Table 2).
| Primer1 | Polarity | Sequence (5’–3’) | Positions (nt) |
| HBV/Ff1 | Sense | GGG TCA CCA TAT TCT TGG GAA | 2 814–2 834 |
| HBV/Fr1 | Antisense | CGT TGC CGA GCA ACG GGG TAA AGG | 1 163–1 140 |
| HBV/Ff2 | Sense | GTT TCT CCT GGC TCA GTT TA | 660–679 |
| HBV/Fr2 | Antisense | AAA AAG TTG CAT GGT GCT GG | 1 825–1 806 |
| HBV/Ff3 | Sense | ACG TCG CAT GGA GAC CAC CG | 1 601–1 620 |
| HBV/Fr3 | Antisense | GAA CTG GAG CCA CCA GCA GG | 75–56 |
| HBV/Ff4 | Sense | CCT CCT GCT TCC ACC AAT CG | 3 124–3 143 |
| HBV/Fr4 | Antisense | AGA TGA GGC ATA GCA GCA GGA TG | 431–409 |
| HBV/Ff5 | Sense | GTC TAG ACT CGT GGT GGA CTT CTC | 246–269 |
| HBV/Fr5 | Antisense | AAG CCA GAC AGT GGG GGA AAG C | 730–709 |
| HBV/Ff6 | Sense | CTC GCC AAC TTA CAA GGC CTT T | 1 098–1 119 |
| HBV/Fr6 | Antisense | GAT TCA GCG CCG ACG GGA CGT A | 1 447–1 426 |
| HBV/Ff7 | Sense | CTC TGC CGA TCC ATA CTG CGG AA | 1 256–1 278 |
| HBV/Fr7 | Antisense | TGA GAT CTT CTG CGA CGC GGC | 2 431–2 410 |
| HBV/Ff8 | Sense | TCA GGC AAG CTA TTT TGT GCT GG | 2 064–2 086 |
| HBV/Fr8 | Antisense | TGT TCA CAT TTG TGT ATC AAA T | 2 587–2 565 |
| HBV/Ff9 | Sense | GCC GCG TCG CAG AAG ATC TCA A | 2 410–2 431 |
| HBV/Fr9 | Antisense | AAA ATG AGG CGC TAT GTG TGG ATT | 2 811–2 788 |
The number of nucleotide substitutions per site was estimated by the 6-parameter method[8], and phylogenetic trees were constructed by the neighbor-joining method[9] based on the numbers of substitutions. To confirm the credibility of phylogenetic analyses, bootstrap resampling tests were carried out 1 000 times[10]. Evolutionary distances between nucleotide sequences of the S gene were calculated in accordance with the HCV Database. These analyses were conducted with use of the ODEN program of the National Institute of Genetics (Mishima, Japan)[11]. Frequency distribution analysis was performed with the StatView J-4.5 program.
Eleven patients with HBV-associated liver disease were found to be infected with HBV/F by ELISA with monoclonal antibodies against preS2 epitopes[5-7]. Table 1 shows demographics and liver disease of the 11 patients from whom HBV/F strains were recovered. Four of them had liver cirrhosis, three had acute hepatitis and one presented with chronic hepatitis; diagnosis of liver disease was not available for the remaining three. They included six Hispanics, two Asians and one Caucasian.
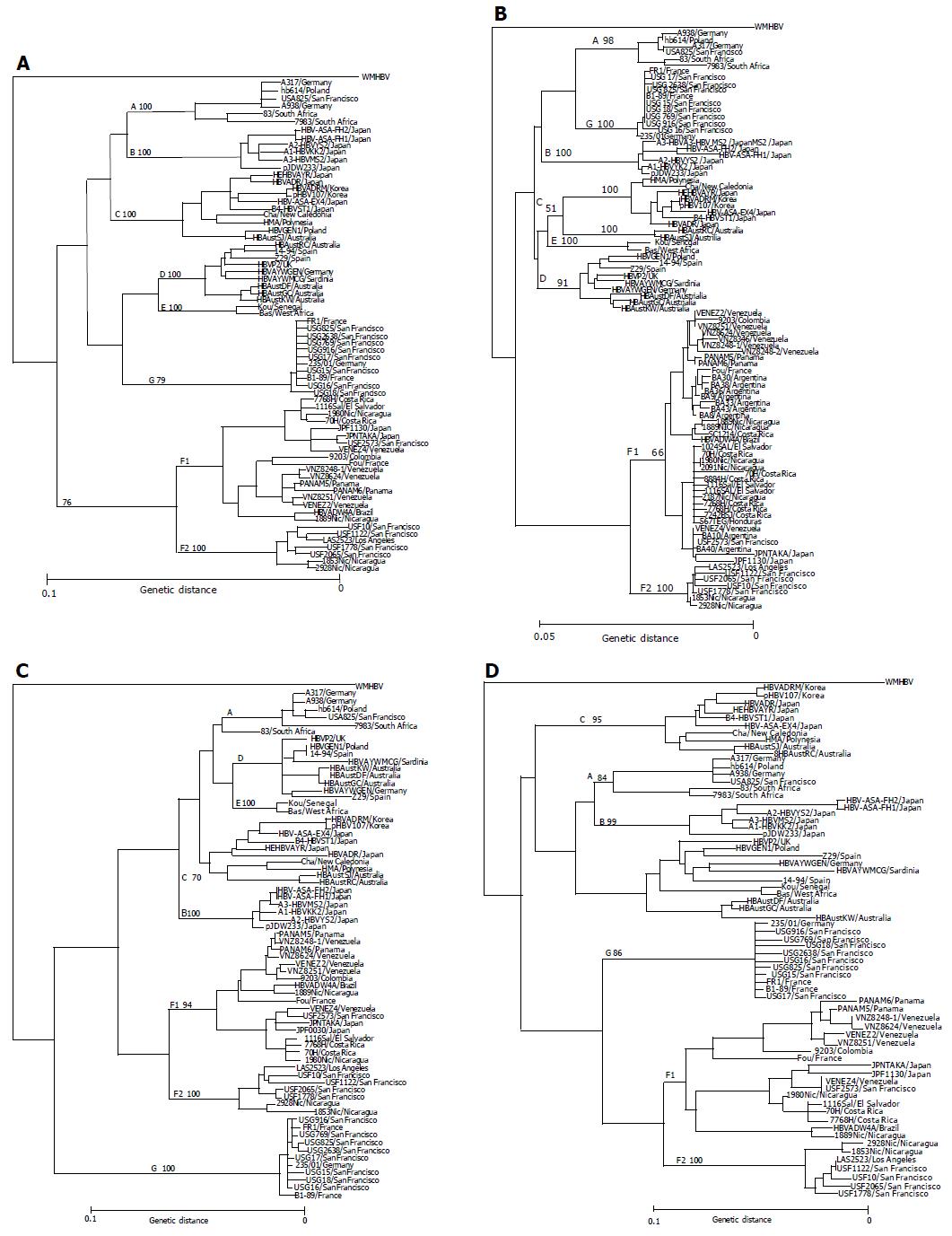
The entire nucleotide sequences of the 11 HBV/F isolates designated USF2573 (accession no. AB064316), USF10 (AB059659), USF1122 (AB064315), USF1778 (AB058660), USF2065 (AB059661), JPF1130 (AB086397), JPNTAKA (AB116654), PAMAN5 (AB116549), PANAM6 (AB116550), VENEZ2 (AB116551) and VENEZ4 (AB116552) have been deposited in the international DDBJ/GenBank/EMBL database. They were subjected to phylogenetic analyses over the entire genome and within the open reading frames among themselves and against 59 HBV isolates of the 7 genotypes (A-G) for which the full-length sequences are known (Figure 1).
Eleven HBV/F and three HBV/H isolates with known entire genomic sequences were recruited from the literature[2,12,13]. One (USF2573) of the five San Franciscan HBV/F isolates, two from Japan (JPF1130 and JPNTAKA), two from Panama (PANAM5 and PANAM6) and two from Venezuela (VENEZ2 and VENEZ4) clustered with the 11 reported HBV/F isolates in phylogenetic trees (Figures 1A-D). The remaining four (USF10, USF1122, USF1778 and USF2065) flocked on a separate branch along with two isolates from Nicaragua and one from Los Angeles, that have been classified into genotype H[4].
The four strains (USF10, USF1122, USF1778 and USF2065) as well as three reported (LAS2523 [accession no. AY090460], 1853Nic [AY090454], 2928Nic [AY090457]) were compared among one another and against the seven isolates each representative of genotype F (AB064316 [USF2573 isolate]) and the other six genotypes (AB064314 for genotype A, D23678 for B, S75184 for C, D23681 for D, X75657 for E and AF160501 for G) over the full-length sequence and regions thereof (Table 3). In the comparison of the full-length sequence, the seven isolates were similar among themselves in ≥96.3%, and to the representative HBV/F isolate in ≥91.0%. They were, however, similar to any representatives of the other six genotypes in only ≤86.5%.
| Genomic regions | Homology within seven HBV/F2 isolates and that of HBV/F2 to seven representative of HBV/F1 as well as genotypes A, B, C, D, E, and G1 | |||||||
| F2 | F1 | A | B | C | D | E | G | |
| Full genome | 97.5 | 91.6 | 86 | 85.9 | 85.7 | 85.7 | 86.1 | 84.5 |
| (96.3–99.2) | (91.0–92.1) | (85.4–86.5) | (85.5–86.2) | (85.2–86.0) | (85.1–86.3) | (85.6–86.4) | (84.1–84.8) | |
| PreS1 region | 98.1 | 90 | 77.3 | 75.7 | 78.9 | 79.3 | 81.4 | 80.6 |
| (97.2–99.4) | (89.1–90.5) | (76.8–78.2) | (75.1–76.5) | (77.9–80.1) | (78.3–80.2) | (80.7–82.0) | (80.4–81.1) | |
| PreS2 region | 97.4 | 90.9 | 82.9 | 79.2 | 82.6 | 81.2 | 83.3 | 81.3 |
| (92.7–100) | (86.1–92.7) | (78.0–84.2) | (75.8–80.6) | (79.4–83.6) | (77.6–82.4) | (79.4–84.2) | (77.0–83.0) | |
| S gene | 99.4 | 96.4 | 92.2 | 92.6 | 91.3 | 92.7 | 92.6 | 92.8 |
| (98.7–99.9) | (95.9–96.8) | (91.8–92.5) | (92.5–92.8) | (91.0–91.6) | (92.4–93.0) | (92.2–92.8) | (92.4–93.0) | |
| C gene | 95.8 | 91.9 | 84.8 | 87.3 | 86.6 | 84.9 | 85.8 | 87.2 |
| (90.9–100) | (88.8–93.5) | (83.0–85.8) | (85.1–88.6) | (84.8–87.5) | (82.8–86.6) | (84.1–86.8) | (84.9–88.2) | |
| P gene | 97.9 | 91.7 | 86.2 | 85.8 | 86.3 | 86.1 | 86.5 | 86.1 |
| (96.6–99.1) | (90.9–92.0) | (85.7–86.6) | (85.4–86.0) | (86.1–86.6) | (85.5–86.4) | (86.2–86.8) | (85.7–86.4) | |
| X gene | 97.1 | 91.1 | 87.7 | 88.7 | 85.4 | 89.2 | 88.3 | 84.2 |
| (94.2–99.8) | (89.3–92.3) | (86.5–88.7) | (86.8–90.1) | (84.7–86.8) | (88.0–90.2) | (87.1–89.5) | (85.0–83.7) | |
Table 4 compares homology in the entire nucleotide sequence among the 11 HBV/F isolates examined in this study and the 13 reported. The sequence divergence among the 7 isolates (USF10, USF1122, USF1778, USF2065, LAS2523, 1853Nic and 2928Nic) and the 10 reported[2,4,12,13] as well as the 7 (USF2573, JPF1130, JPNTAKA, PANAM5, PANAM6, VENEZ2 and VENEZ4) in this study exceeded 7.3%, which fell a little short of clearing a sequence divergence >8% distinguishing the 7 genotypes of HBV[1-3]. It would be appropriate, therefore, to classify the seven isolates into subtypes of genotype F. Thus, F1 subtype (HBV/F1) was assigned to the 11 reported HBV/F isolates and the 7 in this study (USF2573, JPF1130, JPNTAKA, PANAM5, PANAM6, VENEZ2 and VENEZ4) (Figure 1), while F2 subtype (HBV/F2) was consigned to the 7 isolates, 4 from San Francisco (USF10, USF1122, USF1778 and USF2065), 1 from Los Angeles (LAS2523) and 2 from Nicaragua (1853Nic and 2928Nic), all of which were examined in the present study.
| F2 subtype (this study) | F2 subtype (reported) | F1 subtype (this study) | ||||||||||||
| USF10 | USF1122 | USF1778 | USF2065 | 1853Nic | 2928Nic | LAS2523 | USF2573 | JPF1130 | JPNTAKA | PANAM5 | PANAM6 | VENEZ2 | VENEZ4 | |
| HBV/F1 isolates | ||||||||||||||
| (reported) | ||||||||||||||
| AB036905 | 91.7 | 91.9 | 92.3 | 92.6 | 92.3 | 92.7 | 92.5 | 94.6 | 94.1 | 93.6 | 99.2 | 97.9 | 99 | 94.7 |
| AB036910 | 91.7 | 91.9 | 92.3 | 92.7 | 92.2 | 92.6 | 92.6 | 94.7 | 94.1 | 93.6 | 99.2 | 98 | 99.5 | 94.8 |
| AB036920 | 91.7 | 91.9 | 92.2 | 92.5 | 92.2 | 92.7 | 92.5 | 94.6 | 94 | 93.6 | 99.1 | 97.8 | 98.8 | 94.7 |
| X69798 | 91.1 | 91.4 | 91.9 | 92.2 | 91.6 | 92.1 | 92.2 | 94.3 | 93.9 | 93.6 | 96.2 | 95.2 | 96.1 | 94.5 |
| X75659 | 90.5 | 90.7 | 91.2 | 91.6 | 91 | 91.4 | 91.4 | 93.6 | 92.9 | 92.6 | 95.4 | 94.3 | 95.5 | 93.7 |
| X75663 | 91 | 91.2 | 91.8 | 92.2 | 91.7 | 92.1 | 91.9 | 94 | 93.3 | 92.8 | 97.2 | 96 | 97.6 | 94.1 |
| AY090455 | 91 | 91.1 | 91.5 | 91.9 | 91.4 | 91.9 | 91.9 | 94.2 | 93.8 | 93.4 | 96 | 94.9 | 95.8 | 94.3 |
| AY090458 | 90.8 | 90.9 | 91.5 | 92 | 91.6 | 91.9 | 91.6 | 97.9 | 97.3 | 96.9 | 94.9 | 93.8 | 94.9 | 98.1 |
| AY090459 | 90.8 | 90.8 | 91.5 | 91.9 | 91.6 | 91.9 | 91.5 | 98.2 | 97.5 | 97.2 | 95 | 93.9 | 94.9 | 98.3 |
| AY090461 | 90.9 | 91 | 91.6 | 92 | 91.6 | 91.9 | 91.7 | 98.1 | 97.5 | 97.1 | 95 | 93.9 | 94.8 | 98.3 |
| HBV/F1 isolates | ||||||||||||||
| (this study) | ||||||||||||||
| USF2573 | 91 | 91.1 | 91.6 | 92 | 91.8 | 92.1 | 91.8 | – | 98.5 | 97.9 | 94.8 | 93.7 | 94.7 | 99.6 |
| JPF1130 | 90.7 | 90.6 | 91.3 | 91.5 | 91.3 | 91.6 | 91.3 | 98.5 | – | 97.8 | 94.3 | 93.4 | 94.2 | 98.4 |
| JPNTAKA | 90.5 | 90.5 | 91.1 | 91.3 | 91.0 | 91.3 | 91.1 | 97.9 | 97.8 | – | 93.9 | 92.9 | 93.7 | 97.9 |
| PANAM5 | 91.9 | 92 | 92.4 | 92.8 | 92.3 | 92.8 | 92.7 | 94.8 | 94.3 | 93.9 | – | 98.6 | 99.2 | 95 |
| PANAM6 | 91.1 | 91.2 | 91.7 | 91.7 | 91.4 | 91.9 | 91.7 | 93.7 | 93.4 | 92.9 | 98.6 | – | 98 | 93.8 |
| VENEZ2 | 91.8 | 92.1 | 92.5 | 92.8 | 92.3 | 92.8 | 92.7 | 94.7 | 94.2 | 93.7 | 99.2 | 98 | – | 94.9 |
| VENEZ4 | 91.1 | 91.2 | 91.7 | 92.1 | 91.9 | 92.2 | 92 | 99.6 | 98.4 | 97.9 | 95 | 93.8 | 94.9 | – |
| F2 isolates | ||||||||||||||
| (this study) | ||||||||||||||
| USF10 | – | 97 | 96.9 | 97.2 | 96.3 | 96.6 | 97.9 | 91.0 | 90.7 | 90.5 | 91.9 | 91.1 | 91.8 | 91.1 |
| USF1122 | 97 | – | 97.1 | 97.6 | 96.5 | 96.9 | 98.2 | 91.1 | 90.7 | 90.5 | 92 | 91.2 | 92.1 | 91.2 |
| USF1778 | 96.9 | 97.1 | – | 98.6 | 97.2 | 97.4 | 98 | 91.6 | 91.3 | 91.1 | 92.4 | 91.7 | 92.5 | 91.7 |
| USF2065 | 97.2 | 97.6 | 98.6 | – | 97.6 | 97.8 | 98.4 | 92 | 91.5 | 91.3 | 92.8 | 91.7 | 92.8 | 92.1 |
| F2 isolates | ||||||||||||||
| (reported) | ||||||||||||||
| 1853Nic | 96.3 | 96.5 | 97.2 | 97.6 | – | 99.2 | 97.5 | 91.8 | 91.3 | 91.0 | 92.3 | 91.4 | 92.3 | 91.9 |
| 2928Nic | 96.6 | 96.9 | 97.4 | 97.8 | 99.2 | – | 97.7 | 92.1 | 91.6 | 91.3 | 92.8 | 91.9 | 92.8 | 92.2 |
| LAS2523 | 97.9 | 98.2 | 98 | 98.4 | 97.5 | 97.7 | – | 91.8 | 91.3 | 91.1 | 92.7 | 91.7 | 92.7 | 92 |
Three HBV/F2 isolates examined in this study (USF10, USF1778 and USF2065) had the identical genomic length of 3 215 bp as the original three HBV/F isolates including X69798[12], X75658[2] and AB036905[13]. The remaining one HBV/F2 isolate (USF1122) had a shorter length of 3 206 bp due to a deletion of 9 bp in the preS2 region. They all possessed 226 amino acids (aa) coded for by the S gene, 29 aa by the preC region, 183 aa by the C gene, 843 aa by the polymerase (P) gene (840 aa for USF1122) and 154 aa by the X gene. The subtype of HBsAg was deduced to be adw4 in all HBV/F isolates by the expression of Lys122, Leu127, and Lys160[14]. The YMDD motif that is prone to mutation during the lamivudine therapy was identified over aa 549-552 (aa 546-549 in USF1122) with methionine at position 550 (547 in USF1122).
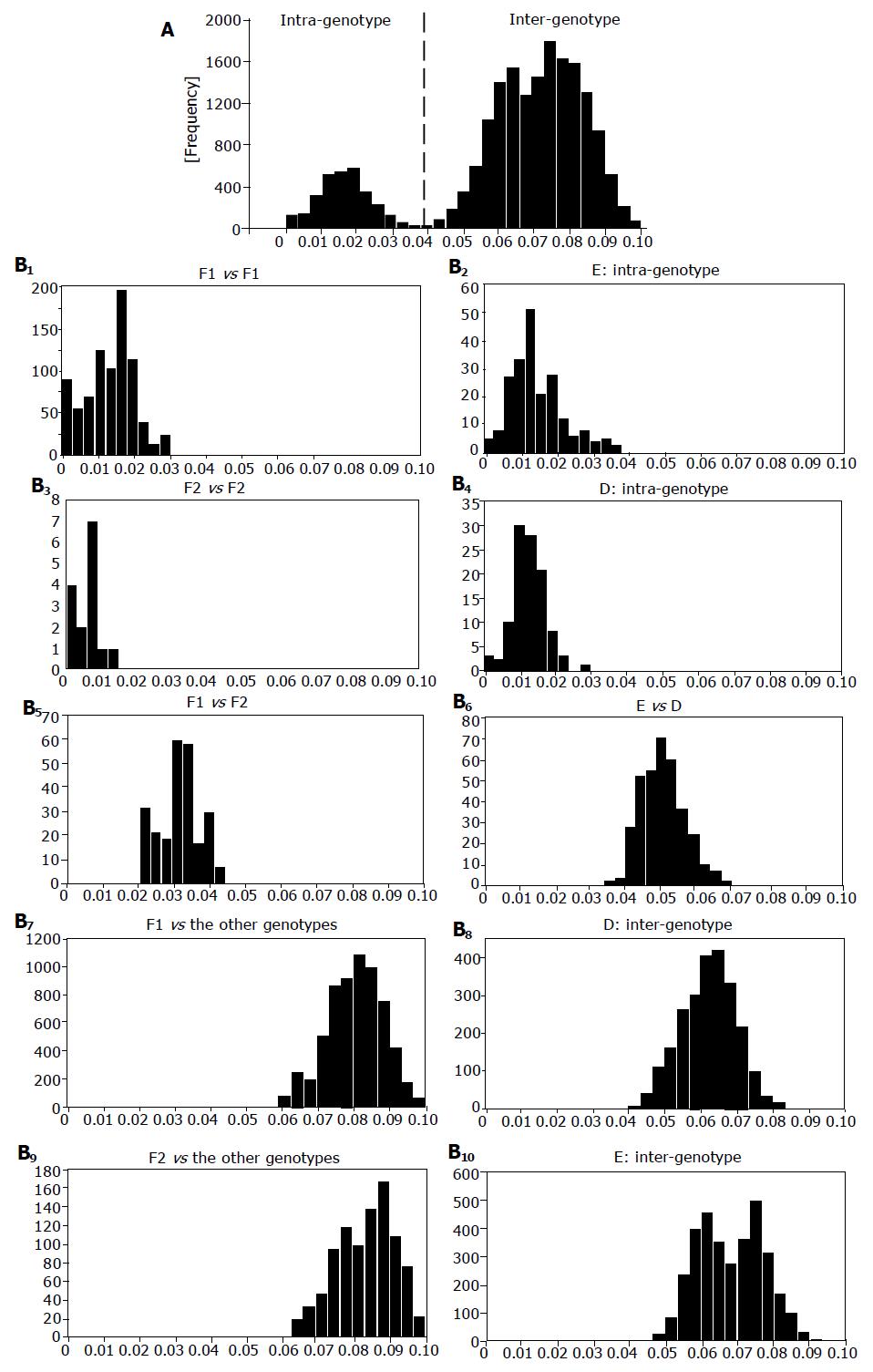
Figure 2 illustrates genetic distance in the S-gene sequences of 47 HBV/F1 isolates, 7 HBV/F2 isolates and 136 HBV isolates of the other 6 genotypes (A-E and G). When all of them were compared two-by-two, they clustered into two heaps that were not separated clearly (Figure 2A). The heap with the mean genetic distance of 0.017 represented divergence of HBV isolates among the same genotype (intra-genotype), and that with the mean distance of 0.073 represented divergence between any two HBV isolates of distinct genotypes (inter-genotype). In Figure 2B, the 7 HBV/F2 and 47 HBV/F1 were compared among themselves (F1 vs F1 or F2 vs F2), each other (F1 vs F2) and against the other genotypes. In this two-by-two comparison, the genetic distance was distributed in two clusters of different orders. The genetic distance among 47 HBV/F1 isolates as well as the 7 HBV/F2 isolates stayed within the intra-genotype heap, and that of HBV/F1 as well as HBV/F2 against other genotypes stood within the inter-genotype heap. Notably, the genetic distance between HBV/F1 and HBV/F2 isolates clustered within the heap of intra-genotype. These distinct hierarchical orders of sequence divergence indicated that HBV/F2 would be closer to HBV/F1 than to any HBV isolates of the other six genotypes (A-E and G). Genetic distance in the S-gene sequences of 22 HBV/E isolates, 16 HBV/D isolates and 158 isolates of the other HBV genotypes is shown in the right half of Figure 2B. The genetic distance within HBV/D and HBV/E stayed within the intra-genotype heap, and that of those against the other genotypes as well as that between HBV/D and HBV/E isolates were kept within the inter-genotype heap.
When all the nucleotide sequences of 68 HBV isolates including 7 HBV/F2 isolates were compared, nucleotide substitutions specific for HBV/F2 were identified at 8 positions (A22, T1467, C1552, G1754, T2495, C2628, C3121, and A3129) (Table 5). Genotype- or subtype-specific nucleotide substitutions clustered within the P gene, and they were unique to each genotype except for genotype A as well as two subtypes of genotype F. Thus, C2606 was specific for genotype B, A2596 for C, C2579/C2634 for D, T2603/C2636 for E, C2558/C2660 for G, T2622 for F1, and C2628 for F2.
| HBV isolates | Nucleotide positions | |||||||||||||||||
| Genotype | P gene | |||||||||||||||||
| /subtype | ||||||||||||||||||
| 22 | 1 467 | 1 552 | 1 754 | 2 495 | 2 558 | 2 579 | 2 596 | 2 603 | 2 606 | 2 622 | 2 628 | 2 634 | 2 636 | 2 660 | 3 121 | 3 129 | ||
| X70185 | A | T | G | T | T | G | T | T | T | C | T | A | A | A | A | T | A | T |
| D00329 | B | – | – | – | – | – | – | – | – | A | C | – | – | – | – | – | G | – |
| D50520 | C | – | – | – | – | A | – | – | A | – | – | – | – | – | – | – | G | – |
| D23681 | D | – | – | – | – | – | A | C | – | – | A | – | – | C | – | – | G | G |
| X75657 | E | – | – | – | – | – | – | – | – | T | – | – | – | – | C | – | G | G |
| AB056515 | G | – | – | – | C | – | C | A | – | – | – | – | – | – | – | C | G | G |
| X68798 | F1 | – | – | – | – | – | – | – | – | – | A | T | – | – | – | – | G | G |
| X75658 | F1 | – | – | – | C | – | – | A | – | – | A | T | – | – | – | – | G | G |
| X75663 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| AB036905 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| AB036910 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| AB036920 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| USF2573 | F1 | – | – | – | – | – | – | A | – | G | – | T | – | – | – | – | G | G |
| JPF1130 | F1 | – | – | – | – | – | – | A | – | – | – | T | – | – | – | – | G | G |
| JPNTAKA | F1 | – | – | – | – | – | – | A | – | – | – | T | – | – | – | – | G | G |
| PANAM5 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| PANAM6 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| VENEZ2 | F1 | – | – | – | C | – | – | A | – | – | G | T | – | – | – | – | G | G |
| VENEZ4 | F1 | – | – | – | T | A | – | A | – | – | – | T | – | – | – | – | G | G |
| USF10 | F2 | A | T | C | G | T | – | A | – | – | A | – | C | – | – | – | C | A |
| USF1122 | F2 | A | T | C | G | T | – | A | – | – | A | – | C | – | – | – | C | A |
| USF1778 | F2 | A | T | C | G | T | – | A | – | – | A | – | C | – | – | – | C | A |
| USF2065 | F2 | A | T | C | G | T | – | A | – | – | A | – | C | – | – | – | C | A |
| AY090454 | F2 | A | T | C | G | T | – | A | – | – | A | G | C | – | – | – | C | A |
| AY090457 | F2 | A | T | C | G | T | – | A | – | – | A | G | C | – | – | – | C | A |
| AY090460 | F2 | A | T | C | G | T | – | A | – | – | A | – | C | – | – | - | C | A |
Substitutions specific for HBV/F2 were identified at the amino acid level within each open reading frame except for the preS2 region. Eight unique amino acid residues were identified within the polymerase (Gln92, Ser267, Thr272, Thr275, Glu282, Thr308, Thr313, and Pro823), four in X (Trp32, Ala60, Pro102 and Leu127), two in preS1 (Ala8 and Pro90), two in HBsAg (Val44 and Pro45) and one in core region (Ala157). Six of the eight (75%) unique amino acid residues identified in the polymerase region were found within the spacer region.
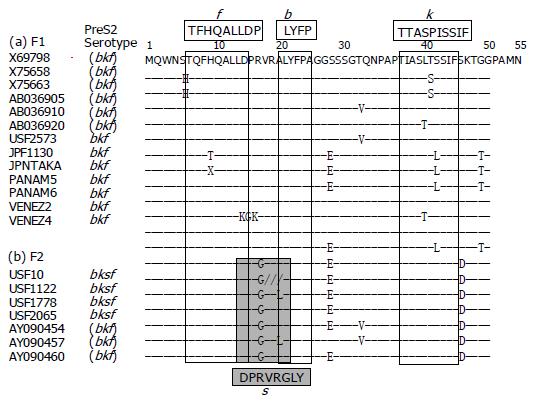
Figure 3 illustrates amino acid sequences of the preS2-region product with reference to the expression of preS2 epitopes detected by four monoclonal antibodies named b, f, k, and s[5]. The preS2 epitope b, which is common to HBV isolates of any genotypes and borne by a tetramer with the sequence of LYFP[5], was not fully conserved in two of the seven HBV/F2 isolates despite the detection of epitope b in their preS2-region products; it was present in all 13 HBV/F1 isolates. The seven HBV/F1 isolates (USF2573, JPF1130, JPNTAKA, PANAM5, PANAM6, VENEZ2, and VENEZ4) had the bkf serotype that was compatible with the preS2 serotype assigned to genotype F[5]. Although serotypes of the six reported HBV/F1 isolates were unknown, they all shared the amino acid sequence for the expression of k and f epitopes; their combination is characteristic of the preS2 serotype (bkf) for genotype F[5].
Unlike the 13 HBV/F1 isolates, all the 7 HBV/F2 isolates possessed G as amino acid 19, and therefore, would bind to the monoclonal antibody against s[5], for the expression of bksf serotype. Thus, the expression of preS2 epitope s would be able to differentiate between F2 and F1 subtypes. Taking the other genotypes into consideration, the combination of preS2 serotype of bksf with HBsAg serotype adw would be specific for HBV/F2, since that with HBsAg serotype adr occurs in HBV isolates of genotype C[5,6]. Hence, HBV/F2 can be serologically differentiated from HBV/F1 as well as the other six genotypes (A-E and G).
Of the seven HBV genotypes (A-G), which are defined by a sequence divergence >8% in the entire genomic sequence, F is unique in that it is the remotest (>15%) from the other six genotypes (A-E and G)[2,12,13]. The origin of HBV/F is proposed in the New World, because it is frequent in Central and South America; F is the major HBV genotype in Venezuela and Argentina[15-19]. A remote genetic distance of genotype F from the other six genotypes mirrors the relationship between HBV of woolly monkeys in the New World and HBV species of major apes (orangutans, gibbons, gorillas, and chimpanzees) in the Old World (reviewed in Ref.[20]). Furthermore, human HBV of genotype F is closer to HBV of woolly monkeys than those of the other six genotypes (A-E and G). Combined, these lines of evidence would point to the co-evolution of human HBV of genotype F and HBV of woolly monkeys in the New World since the distant past, as well as their distinction, respectively, from those of the other six HBV genotypes and HBV species of great apes in the Old World.
Originally, four HBV genotypes (A-D) were proposed by sequence divergence in the entire genome >8%[1]. The four genotypes were followed by three others (E-G) defined by the same criterion[2,3]. In the present study, 11 HBV/F isolates were recovered from patients in San Francisco, Japan, Panama and Venezuela, and they were found to cluster into two groups of four (USF10, USF1122, USF1778 and USF2065) and 7 isolates (USF2573, JPF1130, JPNTAKA, PANAM5, PANAM6, VENEZ2, and VENEZ4) in the phylogenetical analysis on the full-length sequence. The 7 HBV/F isolates in 1 group nested on the same branch as reported 11 HBV/F isolates[2,4,12,13], and the remaining 4 flocked on the same branch as 3 isolates reported under the classification of genotype H[4]. The two groups of HBV isolates differed in the full-length sequence by only >7.3% (range: 9.5-7.3; mean: 8.4±0.6%) (IV).
The observed sequence difference of the seven HBV/F isolates, four in this study (USF10, USF1122, USF1778, and USF2065) and three reported (LAS2523, 1853Nic, and 2928Nic), from the others fell in short of clearing >8% separating the seven HBV genotypes, although two of them (USF10 and USF1122) cleared this margin. It has to be pointed out that the >8% distance was set arbitrarily in 1988 when only a score of HBV isolates with the entire sequence known were available, and before genotype F came along[1]. Although the criterion has helped distinguishing the seven HBV genotypes, it may not be “the gold standard” in genotyping and would need to be modified as new developments arise.
In 1993, Simmonds and his colleagues performed an elegant analysis of HCV isolates by two-by-two comparison of a partial sequence of 222 bp[21]. They found them distribute in three clusters, thereby attesting to the three distinct orders of sequence divergence. They considered a cluster with the shortest evolutionary distance would represent isolate variation, another with a middle distance intra-subtypic variation and the other with the longest distance intra-typic variation. Based on their analysis, they constructed a hierarchical system to classify HCV into six types named by Arabic numbers, each of which breaks down into respective subtypes designated by lower-case alphabet letters. Their classification straightened out a big turmoil created by various nomenclature systems, and has become the standard of HCV genotypes.
The entire sequences of the seven HBV isolates including four in this study (USF10, USF1122, USF1778, and USF2065) and three reported (LAS2523, 1853Nic, and 2928Nic), differing by >7.3% from the other HBV/F isolates[2,12,13], were subjected to the classification method of Simmonds et al[21], within the S-gene sequence (Figure 3). Just like HCV isolates, two clusters with different degrees of genetic distance (intra- and inter-genotypic heap) emerged in the comparison of S-gene sequences of the seven HBV isolates among themselves and against representatives of the seven HBV genotypes (A-G) (Figure 2B). Although the seven HBV isolates were separated from the other HBV/F isolates, they were closer to them than to any HBV isolates of the other six genotypes (A-E and G) in the S-gene sequence.
Bowyer and Sim[22] found ranges of genetic distance among isolates, subtypes and genotypes overlap in the S-gene sequence, thereby preventing distinction between them. In support of their findings, no clear differentiation between isolates and subtypes was observed in pairwise comparison of the S-gene sequence, while genotypes were clearly distinguished from isolates and subtypes (Figure 2A). When looking from HBV/F1 isolates, the genetic distance to HBV/F2 distributed within the intra-genotype range. Hence, it would be justified to classify the four isolates from San Francisco (USF10, USF1122, USF1778, and USF2065) and three reported (LAS2523, 1853Nic, and 2928Nic) into distinct subtypes rather than different genotypes.
On the basis of these analyses, and in hope of avoiding future confusion created by logging into an ever expanding alphabet list of HBV genotypes, four HBV isolates from San Francisco (USF10, USF1122, USF1778, and USF2065) and three reported (LAS2523, 1853Nic, and 2928Nic) would better be classified into subtypes of genotype F, rather than given an independent genotype. Accordingly, the seven variants of HBV/F (USF10, USF1122, USF1778, USF2065, LAS2523, 1853Nic, and 2928Nic) were classified into F2 subtype, while the former HBV/F isolates and the one from San Francisco (USF2573), two from Japan (JPF1130 and JPNTAKA), two from Panama (PANAM5 and PANAM6) and two from Venezuela (VENEZ2 and VENEZ4) into F1 subtype, provisionally. Mbayed et al[18,19], have reported four clusters of HBV/F isolates by comparison of S-gene sequences. F1 subtype covers Clusters I, II and IV, while F2 subtype corresponds to Cluster III in their classification. As the entire genomic sequences of HBV isolates accumulate, there may be HBV/F isolates that are closer to F1 and F2 than to any of the other six genotypes (A-E and G). They will be assigned F3 and F4 subtypes in the chronological order of sequence determination.
There is a possibility for distinguishing between F1 and F2 subtypes by serological means[5,6]. HBV/F2 isolates expressed a preS2 serotype of bksf, in contrast to that of bkf in HBV/F1 isolates. The combination of preS2 serotype of bksf with HBsAg serotype adw appears specific for HBV/F2, distinguishing it from HBV/F1 and the other six genotypes (A-E and G). Serological differentiation of HBV/F2 from HBV/F1 would facilitate their detection in epidemiological and clinical settings. It will allow easy evaluation of F1 and F2 subtypes for different virological virulence and disease-inducing capacity which have been reported for HBV genotypes[23-25].
In classifying microbes, one is tempted to be either a lumper who wants to put groups together in a simplified system or a splitter who likes to break down a clade into many. In classifying HBV genotypes, one may need to lump or split them as required, taking into considerations virological phylogeny and clinical relevance of each genotype or subtype[26,27]. For instance, there are two subtypes of genotype B, one of which has the recombination with genotype C in the precore region and core gene, while the other does not have it[28]. Because these two subtypes of genotype B influence the response to lamivudine in patients with chronic hepatitis B, they are clinically useful[29]. Our proposal for two subtypes of HBV genotype F based on phylogenetic analysis would be preferred to classifying them into genotypes F and H[4]. The method for serologically distinguishing between subtypes F1 and F2 would facilitate the evaluation of their differences, some of which may turn out to be clinically relevant.
Dr. Masashi Mizokami was granted by Ministry of Health, Labour and Welfare of Japan (H13-kaken-2).
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence, comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575-2583. [DOI] [Full Text] |
| 2. | Norder H, Couroucé AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 556] [Cited by in RCA: 587] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 3. | Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67-74. [PubMed] |
| 4. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. [PubMed] |
| 5. | Usuda S, Okamoto H, Iwanari H, Baba K, Tsuda F, Miyakawa Y, Mayumi M. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods. 1999;80:97-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 224] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 6. | Usuda S, Okamoto H, Tanaka T, Kidd-Ljunggren K, Holland PV, Miyakawa Y, Mayumi M. Differentiation of hepatitis B virus genotypes D and E by ELISA using monoclonal antibodies to epitopes on the preS2-region product. J Virol Methods. 2000;87:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Kato H, Orito E, Sugauchi F, Ueda R, Gish RG, Usuda S, Miyakawa Y, Mizokami M. Determination of hepatitis B virus genotype G by polymerase chain reaction with hemi-nested primers. J Virol Methods. 2001;98:153-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Gojobori T, Ishii K, Nei M. Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol. 1982;18:414-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 179] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-425. [PubMed] |
| 10. | Felsenstein J. Confidence limits on phylogenesis, an approach using the bootstrap. Evolution. 1985;39:783-791. [RCA] [DOI] [Full Text] [Cited by in Crossref: 13218] [Cited by in RCA: 13370] [Article Influence: 334.3] [Reference Citation Analysis (0)] |
| 11. | Ina Y. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput Appl Biosci. 1994;10:11-12. [PubMed] |
| 12. | Naumann H, Schaefer S, Yoshida CF, Gaspar AM, Repp R, Gerlich WH. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J Gen Virol. 1993;74:1627-1632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 91] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Nakano T, Lu L, Hu X, Mizokami M, Orito E, Shapiro C, Hadler S, Robertson B. Characterization of hepatitis B virus genotypes among Yucpa Indians in Venezuela. J Gen Virol. 2001;82:359-365. [PubMed] |
| 14. | Okamoto H, Imai M, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Point mutation in the S gene of hepatitis B virus for a d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J Virol. 1987;61:3030-3034. [PubMed] |
| 15. | Arauz-Ruiz P, Norder H, Visoná KA, Magnius LO. Genotype F prevails in HBV infected patients of hispanic origin in Central America and may carry the precore stop mutant. J Med Virol. 1997;51:305-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Arauz-Ruiz P, Norder H, Visoná KA, Magnius LO. Molecular epidemiology of hepatitis B virus in Central America reflected in the genetic variability of the small S gene. J Infect Dis. 1997;176:851-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Blitz L, Pujol FH, Swenson PD, Porto L, Atencio R, Araujo M, Costa L, Monsalve DC, Torres JR, Fields HA. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J Clin Microbiol. 1998;36:648-651. [PubMed] |
| 18. | Mbayed VA, López JL, Telenta PF, Palacios G, Badía I, Ferro A, Galoppo C, Campos R. Distribution of hepatitis B virus genotypes in two different pediatric populations from Argentina. J Clin Microbiol. 1998;36:3362-3365. [PubMed] |
| 19. | Mbayed VA, Barbini L, López JL, Campos RH. Phylogenetic analysis of the hepatitis B virus (HBV) genotype F including Argentine isolates. Arch Virol. 2001;146:1803-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Robertson BH, Margolis HS. Primate hepatitis B viruses - genetic diversity, geography and evolution. Rev Med Virol. 2002;12:133-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391-2399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 928] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 22. | Bowyer SM, Sim JG. Relationships within and between genotypes of hepatitis B virus at points across the genome: footprints of recombination in certain isolates. J Gen Virol. 2000;81:379-392. [PubMed] |
| 23. | Mayerat C, Mantegani A, Frei PC. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J Viral Hepat. 1999;6:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 201] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 24. | Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 699] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 25. | Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 315] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 26. | Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 224] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 27. | Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Gish RG. Epidemiologic and virologic characteristics of hepatitis B virus genotype B having the recombination with genotype C. Gastroenterology. 2003;124:925-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 164] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 28. | Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985-5992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 233] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 29. | Akuta N, Suzuki F, Kobayashi M, Tsubota A, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Arase Y. The influence of hepatitis B virus genotype on the development of lamivudine resistance during long-term treatment. J Hepatol. 2003;38:315-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |