Published online Oct 21, 2005. doi: 10.3748/wjg.v11.i39.6130
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: October 21, 2005
AIM: To evaluate the activity of apoptosis in liver tissue and explore its possible association with hepatic necroinflam-mation and fibrosis as well as serum hepatitis C virus (HCV) load.
METHODS: The studied population included 50 chronic hepatitis C patients (20 women and 30 men, aged 18-66 years). HCV-RNA quantification was performed by two-step real-time quantitative RT-PCR method using the TaqMan technology (reagents of Applera Corporation firm, USA). The morphology of liver tissue was assessed descriptively and scored (necroinflammatory activity and fibrosis). The early apoptosis activity in liver tissue was examined by ssDNA apoptosis ELISA kit, (Chemicon, Germany).
RESULTS: The correlation between apoptosis and fibrosis in liver tissue was observed. High intensification of apoptosis was proportional to the increase of fibrosis (ssDNA: 16.65×10-5 mg/g; 12.71×10-5 mg/g), however, this difference was not statistically significant (P>0.05). Activity of apoptosis in the liver tissue, expressed by ssDNA concentration did not depend on hepatic necroinflammatory changes, HCV-RNA viral load, ALT, and AST activity as well as prothrombin time and INR index.
CONCLUSION: Fibrosis in the tissue is closely associated with early apoptosis in HCV-infected patients.
- Citation: Lapiński TW, Panasiuk A, Jaroszewicz J, Kowalczuk O, Flisiak R, Rogalska M. Specific ssDNA concentration in liver tissue as an index of apoptosis in hepatitis C virus-infected patients. World J Gastroenterol 2005; 11(39): 6130-6133
- URL: https://www.wjgnet.com/1007-9327/full/v11/i39/6130.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i39.6130
Activity of apoptosis during hepatitis C virus (HCV) infection is a result of inhibitory and stimulatory effects of viral proteins. Apoptosis of HCV-infected hepatocytes influences the elimination of viruses. Insufficient activity of this process can lead to persistent liver inflammation[1]. On the contrary, excessive activity of apoptosis causes uncontrolled damage to liver cells. Moreover, there is evidence that apoptosis influences fibrosis progression.
This study was to evaluate the activity of apoptosis in liver tissue and explore its possible association with hepatic necroinflammation and fibrosis activity as well as serum HCV viral load.
The study included 50 chronic hepatitis C patients (20 women and 30 men, aged 18-66 years). Their HCV infection did not exceed 5 years. Inclusion criteria were the following: no focal changes in ultrasonography investigation of liver, absence of drugs or alcohol abuse, autoimmune disease, HIV and hepatitis B co-infection as well as neoplasmatic and other serious diseases, which might alter apoptosis activity. Informed consent was obtained from each patient and the Bioethics Committee at the Medical University of Bialystok approved the study protocol.
The presence of anti-HCV antibodies in serum was detected by MEIA (Abbott, USA). Test was based on recombinant core proteins for HCr43, structural proteins for c200 (NS3 and NS4) and unstructured proteins for c100-3 (NS4) and NS5.
Blood samples were obtained from 47 patients with known chronic hepatitis C. All blood samples were collected to Vacutainer tubes with no additives and centrifuged within 2 h of collection. The serum was aliquoted and kept at -80 °C until further use.
RNA was extracted from 280 mL of each serum sample using Qiagen (Hilden, Germany) QIAamp viral RNA mini kit according to the manufacturer's protocol and dissolved in 50 mL of RNase-free water. This resulted in nearly sixfold concentration of the original output material. RNA was stored at -80 °C until further testing. Each isolation was carried out in duplicate.
HCV-RNA quantification was performed by two-step real-time quantitative RT-PCR method using the TaqMan technology[2]. First, 3.5 mL of total RNA was subjected to reverse transcription in 10 mL of reaction mixture using the Applied Biosystems (Applera Corporation, USA) TaqMan reverse transcription reagents and random hexamers (dN)6. RNA was previously denatured at 70 °C for 10 min. The reaction mixture was prepared as follows: 1×reverse transcription buffer, 5.5 mmol/L MgCl2, 0.5 mmol/L of each dNTPs, 2.5 mmol/L of primer (dN)6, 0.4 U of RNase inhibitor per mL and 1.25 U of MultiScribe transcriptase per mL. The reaction was carried out at 48 °C for 30 min preceded by the incubation at 25 °C for 10 min. Incubation at 95 °C for 5 min terminated the reaction. The resulting cDNA was used as a template for PCR amplification.
cDNA amplification was performed by the Applied Biosystem (Applera Corporation, USA) ABI Prism 7 900HT sequence detection system in TaqMan universal PCR master mix, which is optimized for TaqMan reactions and contains Applied Biosystems (Applera Corporation, USA) AmpliTaq gold DNA polymerase, AmpErase UNG, dNTPs with UTP, passive reference ROX and optimized buffer components. For each PCR reaction, 4 mL of cDNA was added to 16 mL of PCR mix containing 450 nmoL each of HCV-specific forward and reverse primers and 300 nmoL of HCV-specific FAM-MGB probe. The primers and probe were designed and manufactured by Applied Biosystems (Applera Corporation, USA) and optimized for HCV genotype RNA detection. Thermal cycling conditions were designed as follows: inactivation of possible contaminating amplicons by AmpErase UNG at 50 °C for 2 min, initial cDNA denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 60 s. All amplification reactions were carried out in triplicate.
Fluorescent measurements were recorded during the annealing step of each cycle. At the end of each PCR run, data were automatically analyzed by the system to generate the amplification plots and the threshold cycle (Ct) for each sample was calculated. Sample RNA concentrations were then automatically calculated by interpolation of the exper-imentally determined standard curve.
An external standard curve was generated with each run by amplification of 10-fold serial dilutions of RNA isolated from the viral quality control serum (PeliSpy, Sanguin), containing 38 000 geq/mL HCV. RNA isolation and cDNA synthesis were carried out as described for unknown samples. All amplification reactions were carried out in duplicate. The standard curve was created automatically by the ABI Prism 7 900HT detection system by plotting the Ct values against each standard dilution of known concentration.
Precautions were undertaken to minimize the risk of PCR contamination during analysis.
Liver tissue was obtained from the right lobe of the liver (with 1.6-1.8 mm needles from Hypafix packs by Braun, Germany). A part of the liver tissue was subjected to morphologic examination. The morphology of liver tissue was assessed descriptively and scored (necroinflammatory activity and fibrosis) according to Scheuer[3].
Another part of the liver tissue was transferred to 0.9% NaCl buffer and then subjected to apoptosis activity measurement.
The early apoptosis activity in liver tissue was measured by ssDNA apoptosis ELISA kit, (Chemicon, Germany) based on the mAb to single-stranded DNA (ssDNA). The intensity of the reaction between ssDNA and mAb was evaluated. The amount of ssDNA was calculated with reference to absorbance line of positive and negative control.
The liver tissue was homogenized in 0.9% NaCl buffer. The concentrations of ssDNA and total proteins in the homogenized liver tissue were analyzed in duplicate. The homogenates of liver tissue were transferred to wells 24 h prior to the succeeding steps of the procedure to allow cell attachment. Then formamide was added to denature DNA in apoptotic cells, but not in necrotic cells or in cells with DNA damage in the absence of apoptosis. This process enabled specific receptors of apoptotic ssDNA binding to mAb. Following this reaction, 3-ethylbenziazoline-6-sulfonic acid was added to color the products of this reaction[4].
Absorbance was defined at 405 nm. The concentration of ssDNA was calculated from a prepared standard curve of absorbance (basis of different increasing positive and negative control). The results of ssDNA were referred to 1.0 g of proteins in the liver tissue sample.
Statistical analysis was performed by non-parametric Mann-Whitney U, Wilcoxon rank, and Pearson tests. P<0.05 was considered statistically significant.
Diagnostic biopsy was obtained from 47 patients. In three of the remaining cases, the liver biopsy sample did not contain five portobiliary areas, therefore it was impossible to obtain reliable histology results.
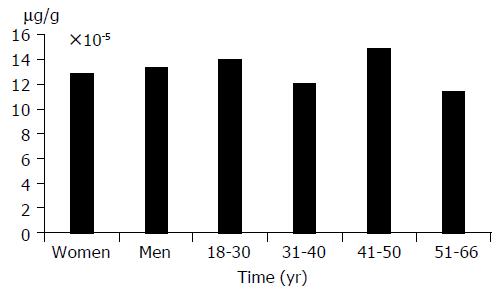
The activity of apoptosis in liver tissue of the studied group was independent of sex and age. ssDNA concentration in liver tissue did not correlate with ALT, AST activity, prothrombin time, and INR index. Moreover, apoptosis was independent of necroinflammatory changes in liver tissue (Table 1).
| ALT | AST | Prothrombintime | INRindex | |
| Pearson test | 0.08 | 0.14 | -0.01 | 0.03 |
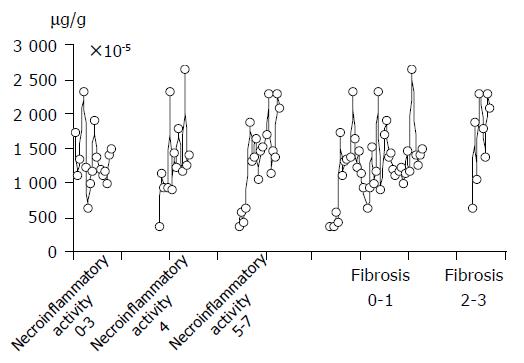
The correlation between fibrosis and apoptosis was observed. Concentration of ssDNA was higher (16.65×10-5 mg/g) in patients with more intensified fibrosis of liver tissue than in those with fibrosis (12.71×10-5 mg/g). However, this difference was not statistically significant (P>0.05). (Figures 1 and 2)
The serum HCV-RNA concentration did not correlate with ALT, AST activity, prothrombin time, and INR index. Moreover, no correlation between serum HCV-RNA and ssDNA concentrations in liver tissue was observed (Table 2).
| n | G1+G2 | Concentration ofssDNA (μg/g) | Concentrationof HCV-RNA(geq/mL) | Pearson test | n | S | Concentration of ssDNA (μg/g) | Concentrationof HCV-RNA (geq/mL) | Pearsontest |
| 16 | 0-3 | 13.16×10-5 | 4.96-54.2×103 | -0.11 | 39 | 0-1 | 12.71×10-5 | 4.96-227×103 | 0.01 |
| 3.95×10-5 SD± | 4.85-10-5 SD± | ||||||||
| 13 | 4 | 13.41×10-5 | 2.48×10-227×103 | -0.14 | 8 | 2-3 | 16.65×10-5 | 14.7-266×103 | 0.01 |
| 6.0×10-5 SD±; | 5.98×10-5 SD± | ||||||||
| 18 | 5-7 | 13.56×10-5 | 14.7-266×103 | -0.13 | |||||
| 5.85×10-5 SD±; |
Apoptosis is one of the factors regulating the course of chronic viral hepatitis. Since this process runs gradually, modification of its course is possible. Different HCV proteins can accelerate or suppress apoptosis.
HCV core proteins overactivate Fas particles expressed on hepatocytes, leading to degradation of cell DNA[5,6]. The Fas particles are "death domains"and indicators of apoptosis. Higher Fas concentration in patients with HCV infection can increase the activity of apoptosis.
HCV non-structural proteins (NS5A) are able to suppress apoptosis by diminishing synthesis of caspase 3, protein kinase A and suppressing TNF-a stimulating polymerase[7,8]. Insuf-ficient activity of apoptosis can influence the persistence of chronic inflammation related to HCV infection[1].
Di Martino et al[9] showed that activity of apoptosis in chronic hepatitis C patients is proportional to HCV-RNA levels. Caronia et al[10] have shown that HCV strongly inhibits apoptosis. In previous investigations, the concentration of ssDNA, as an early indicator of apoptosis was not correlated with HCV viral load, functional state of the liver and necroinflammatory changes in liver tissue samples.
Tsamandas et al[11] evaluated the location and concentration of Bcl-2 and Bax protein, markers of apoptosis activity, in patents with HBV and HCV infection, and found that Bcl-2 and Bax concentrations are enhanced in the regions of intense fibrosis. Takehara et al[12] have shown the linear dependence between the concentration of Bcl-2 family members and activity of fibrosis in the liver. Our results are in accordance with their observations.
Bcl-2 and Bax proteins can mark the late period of apoptosis. Correlation between apoptosis and fibrosis in liver tissue seems unquestionable. However, whether cells with high activity of apoptosis are molecular inductors of fibrosis in HCV-infected patients remains to be solved.
In conclusion, fibrosis of liver tissue is associated with HCV-infection, but apoptosis is not associated with necroinflammatory changes in chronic hepatitis C patients.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Zuckerman E, Zuckerman T, Sahar D, Streichman S, Attias D, Sabo E, Yeshurun D, Rowe J. bcl-2 and immunoglobulin gene rearrangement in patients with hepatitis C virus infection. Br J Haematol. 2001;112:364-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | Castelain S, Descamps V, Thibault V, François C, Bonte D, Morel V, Izopet J, Capron D, Zawadzki P, Duverlie G. TaqMan amplification system with an internal positive control for HCV RNA quantitation. J Clin Virol. 2004;31:227-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 65] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 3. | Scheuer PJ. Classification of chronic viral hepatitis: a need for reassessment. J Hepatol. 1991;13:372-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1130] [Cited by in RCA: 1197] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 4. | Frankfurt OS, Krishan A. Apoptosis enzyme-linked immunosorbent assay distinguishes anticancer drugs from toxic chemicals and predicts drug synergism. Chem Biol Interact. 2003;145:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | Hahn CS, Cho YG, Kang BS, Lester IM, Hahn YS. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology. 2000;276:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 95] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Zhu N, Ware CF, Lai MM. Hepatitis C virus core protein enhances FADD-mediated apoptosis and suppresses TRADD signaling of tumor necrosis factor receptor. Virology. 2001;283:178-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 75] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Ezelle HJ, Balachandran S, Sicheri F, Polyak SJ, Barber GN. Analyzing the mechanisms of interferon-induced apoptosis using CrmA and hepatitis C virus NS5A. Virology. 2001;281:124-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Di Martino V, Brenot C, Samuel D, Saurini F, Paradis V, Reynes M, Bismuth H, Feray C. Influence of liver hepatitis Cvirus RNA and hepatitis C virus genotype on Fas-mediated apoptosis after liver transplantation for hepatitis C. Transplantation. 2000;15:1390-1396. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Caronia S, McGarvey MJ, Goldin RD, Foster GR. Negative correlation between intrahepatic expression of hepatitis C antigens and apoptosis despite high-level expression of Fas and HLA antigens. J Viral Hepat. 2004;11:511-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Tsamandas AC, Thomopoulos K, Zolota V, Kourelis T, Karatzas T, Ravazoula P, Tepetes K, Petsas T, Karavias D, Karatza C. Potential role of bcl-2 and bax mRNA and protein expression in chronic hepatitis type B and C: a clinicopathologic study. Mod Pathol. 2003;16:1273-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Takehara T, Tatsumi T, Suzuki T, Rucker EB, Hennighausen L, Jinushi M, Miyagi T, Kanazawa Y, Hayashi N. Hepatocyte-specific disruption of Bcl-xL leads to continuous hepatocyte apoptosis and liver fibrotic responses. Gastroenterology. 2004;127:1189-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 195] [Article Influence: 9.3] [Reference Citation Analysis (0)] |