Published online Oct 14, 2005. doi: 10.3748/wjg.v11.i38.5926
Revised: April 10, 2004
Accepted: April 13, 2004
Published online: October 14, 2005
AIM: To characterize the expression and genomic amplification of decoy receptor 3 (DcR3) in hepatocellular carcinoma (HCC) and to evaluate the role of DcR3 in apoptosis.
METHODS: We examined 48 cases of HCC for DcR3 expression by RT-PCR and DcR3 gene amplification by quantitative genomic PCR. DcR3 protein was detected by immunohistochemistry. Terminal deoxynucleotidyl transferase-mediated dUTP digoxigenin nick and labeling (TUNEL) was used to identify the apoptosis cells in tissues. Primary hepatoma cell culture and MTT test were used to evaluate the protection against FasL- and chemical-induced apoptosis by DcR3 expression.
RESULTS: DcR3 mRNA overexpression was detected in 60% HCC (29/48) patients. The occurrence of HCC was not associated with amplification of the gene. One sample base substitution was found in three sites as a sequence in Genbank. The expression of DcR3 in HCC was associated with the apoptotic index (0.067±0.04 vs 0.209±0.12, P<0.01), size of mass, stage, and infiltration or metastasis (41.2% vs 71.0%, 40% vs 75%, 51.8% vs 84.6%, P<0.05). DcR3 expression could protect hepatoma cells against apoptosis induced by FasL, but not by chemicals.
CONCLUSION: These data suggest that in addition to gene amplification there may be another mechanism underlying DcR3 overexpression. The effect of overexpression of DcR3 on the apoptosis of cancer cells may have direct therapeutic implications for the management of HCC.
- Citation: Shen HW, Gao SL, Wu YL, Peng SY. Overexpression of decoy receptor 3 in hepatocellular carcinoma and its association with resistance to Fas ligand-mediated apoptosis. World J Gastroenterol 2005; 11(38): 5926-5930
- URL: https://www.wjgnet.com/1007-9327/full/v11/i38/5926.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i38.5926
DcR3, a member of the tumor necrosis factor receptor superfamily, is amplified and overexpressed in various human cancers as a negative regulator of Fas-mediated apoptosis[1-3]. It was reported that DcR3 can bind to LIGHT and FasL[4-8]. Therefore, DcR3 may act as an inhibitor of LIGHT-induced tumor cell death by blocking LIGHT interaction with its receptors. DcR3 is characterized by a soluble cognate receptor for both LIGHT and FasL/CD95L[9,10]. LIGHT and FasL mediate apoptosis, which is the most common physiological form of cell death and occurs during embryonic development, tissue remodeling, immune regulation, and tumor regression. Abnormalities in FasL and LIGHT system can result in a number of human pathological conditions such as tumor. The cause of DcR3 overexpression in human cancers remains unclear, but its value in tumor escaping from apoptosis and immune surveillance of organism is worthy of special remark.
In contrast, no data are available regarding the expression and amplification of DcR3 gene in HCC. We found that there was a high DcR3 protein level in serum of patients with HCC. In the present study, we examined 48 cases of HCC for DcR3 gene expression by RT-PCR and DcR3 gene amplification by quantitative genomic PCR. The expression of DcR3 protein was detected by immunohistochemistry. In situ TUNEL was used to identify the apoptosis cells in tissues of HCC. Primary hepatoma cell culture and MTT test were used to evaluate the role of DcR3 expression in protection against FasL and chemical-induced cell death. The findings in our study suggest that in addition to gene amplification there may be another mechanism underlying DcR3 gene expression. The effect of overexpression of DcR3 on the apoptosis of cancer cells may have direct therapeutic implications for the management of HCC, which remains a major unsolved health problem throughout the world.
Quantitative genomic PCR was performed as previously described[2,11]. Genomic DNA was isolated from fresh tissues of HCC. Quantitative PCR was carried out by a lightcycler. Primers for DcR3 were designed using an intron sequence to avoid amplification from DcR3 mRNA. The DcR3-specific primers were 5’CTTCTTCGCGCACGCTG-3’(sense) and 5’ATCACGCCGGCACCAG-3’(antisense), and the fluorogenic probe was 5’FAM-ACACGATGCGTGC-TCCAAGCAGAA-TAMARA-3’ The beta-globin primers were 5’ACCCTTAGGCTGCTGGTGG-3’(sense) and 5’GGAGTGGACAGATCCCCAAA-3’(antisense), and the fluorogenic probe was 5’FAM-CTACCCTTGGACCCA-GAGGTTCTTTGAGTC-TAMARA-3’ For each run, DNA isolated from normal matched tissues was used for comparison.
For RT-PCR analysis, total RNA was isolated using Trizol (Life Technologies, Inc.) from the tumor and its adjacent normal tissues of 48 HCC cases. RNA was converted to cDNA by reverse transcription and amplified for 35 cycles by PCR. Primers used for amplification of the DcR3 fragment were according to the RNA sequence of DcR3. Beta-actin was used as an internal control for RNA integrity. The DcR3-specific primers were 5’TCCACCGCGCCACTACAC-3’(sense) and 5’ACGGCACGCTCACACTCC-3’(antisense). The beta-actin specific primers were 5’TTCCAGCCT-TCCTTCCTGG-3’(sense) and 5’TTGCGCTCAGG-AGGAGCA AT-3’(antisense). PCR products were run on 2% agarose gel, stained with ethidium bromide, and visualized by UV illumination. RT-PCR product of one sample was sequenced to show reliability of the reaction.
HCC tissues were obtained from the Surgical Department of the Second Affiliated Hospital of Zhejiang University. Four micrometer-thick formalin-fixed and paraffin-embedded sections were deparaffinized and treated in microwave with citrate buffer, pH 6.0 (three times each for 5 min) for antigen retrieval and incubated for 15 min in 30 mL/L H2O2 Tris-buffered saline (TBS), pH 7.4 to block endogenous peroxidase activities. The sections were then incubated for 10 min at room temperature with blocking solution. Each section was incubated with anti-DcR3 antibody diluted 1:5000 for 30-60 min at room temperature. Bound antibodies were detected with the peroxidase-labelled streptavidin-biotin method, and stained with diaminobenzidine. Counterstaining was performed with Mayer’s heamatoxylin, and the sections were mounted.
As a negative control for DcR3 protein expression, the primary antibody was replaced by normal non-immune rabbit serum. There were no known normal tissues expressing DcR3 protein, and no positive control was set up in the DcR3 immunostaining.
HCC tissues were obtained from the Surgical Department of the Second Affiliated Hospital of Zhejiang University. Four micrometer-thick formalin-fixed and paraffin-embedded sections were deparaffinized and treated in microwave with citrate buffer, pH 6.0 (three times each for 5 min) for antigen retrieval and incubated for 15 min in 30 mL/L H2O2 TBS, pH 7.4 to block endogenous peroxidase activities. Each section was incubated with TUNEL reaction solution (Roche) for 60 min at 37 °C, and washed three times with PBS. The signal translation solution was added to the slides for 30 min at
37 °C After being washed three times with PBS, each section was stained with freshly prepared diaminobenzidine. Counterstaining was performed with Mayer’s heamatoxylin, and the sections were mounted.
As a negative control for in situ TUNEL, the TUNEL reaction solution was replaced by TBS. As a positive control, a slide (Roche) in the kit was used together with the samples.FasL-induced apoptosis and drug-induced cell death of hepatoma cells
Hepatoma cells were obtained from patients with HCC who underwent surgery for tumor resection. After tumor removal, the tissues were placed immediately in petri dishes, minced mechanically, and digested by collagenase (3 h, 37?). Subsequently, the dissociated cells were filtered through 100 mm of cell strainers to remove tissue debris. After centrifugation and lysis of erythrocytes by treatment with hypotonic water, the hepatoma cells were washed and resuspended in full medium (CAM).
The hepatoma cells were cultured with serum-free CAM in 96-well plates. A total 5×104 cells were seeded per well. Then 3 ng/mL FasL and ADM, MMC, and GEMZAR of 4-, 1- and 0.25-fold drug concentrations respectively were used to detect the cytotoxic effect on primary hepatoma cells. Each concentration and blank control were put in three parallel wells. The hepatoma cells were co-cultured with FasL or drugs in serum-free CAM for 5 d. The survival rate of the cells was detected with MTT method.
Results were expressed as mean±SE. Differences between groups were examined for statistical significance using analysis of variance (ANOVA) and t-test. P<0.05 was considered statistically significant.
We report that DcR3 mRNA is overexpressed in a substantial percentage of HCC, and that this overexpression can occur in the absence of detectable DcR3 gene amplification. In a case of sequence analysis, three base differences in the DcR3 gene of HCC were identified. The expression of DcR3 in HCC shows a relationship with the apoptosis index of the tissues of cancer. We also demonstrate that the hepatoma cells expressed DcR3 are protected from FasL-induced apoptotic cell death, but not the cell death induced by chemicals.
The mean age of cancer patients was 58.3±14 years. The clinical and pathological features of the total 48 cases were collected and summarized. The relationship between the DcR3 mRNA expression and the clinical and pathological features was analyzed. DcR3 mRNA expression in HCC showed a relationship with size of the mass, TNM stage and infiltration or metastasis of the tumor (P<0.05), but no relationship with membrane of the tumor, cancer embolus, and number of the tumor nodes (Table 1).
| Features | n | DcR3 mRNA expression(%) | p |
| Size of mass | |||
| ≤ 5 cm | 17 | 41.2 (7/17) | <0.05 |
| > 5 cm | 31 | 71.0 (22/31) | |
| Number of node | |||
| Single | 32 | 53.1 (17/32) | >0.05 |
| Multiple | 16 | 75 (12/16) | |
| Membrane of the mass | |||
| Intact | 14 | 50 (7/14) | >0.05 |
| Not intact | 34 | 64.7 (22/34) | |
| Cancer embolus | |||
| Negative | 38 | 60.5 (23/38) | >0.05 |
| Positive | 10 | 60 (6/10) | |
| TNM stage | |||
| Stage I,II | 20 | 40 (8/20) | <0.05 |
| Stage III,IV | 28 | 75 (21/28) | |
| Infiltration or metastasis | |||
| Negative | 35 | 51.4 (18/35) | <0.05 |
| Positive | 13 | 84.6 (11/13) |
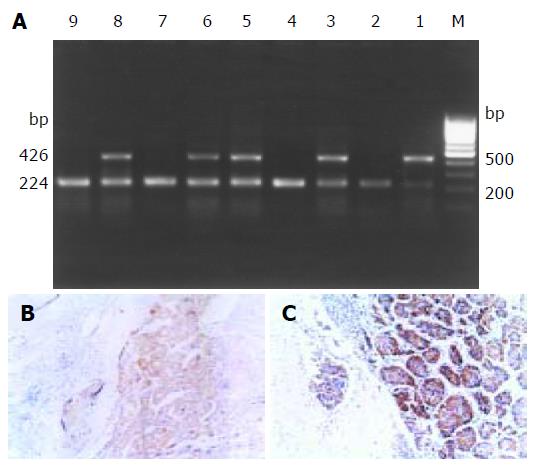
Twenty-nine of 48 HCC cases were positive for DcR3 mRNA expression (Figure 1A). The ratio of positive expression was 60.4% (29/48). No positive expression was detected in adjacent normal tissues. As the level of DcR3 mRNA appeared to be elevated in HCC, we investigated whether it was true for DcR3 at the protein level. The expression of DcR3 protein was detected in five cases of HCC by immunohistochemistry. An obvious positive staining signal of DcR3 protein was detected in three out of five HCC cases (Figures 1B, and C). RT-PCR showed positive signals only in cancer cells. Furthermore, we found a high DcR3 protein level in serum of HCC patients by ELISA with anti-DcR3 antibody.
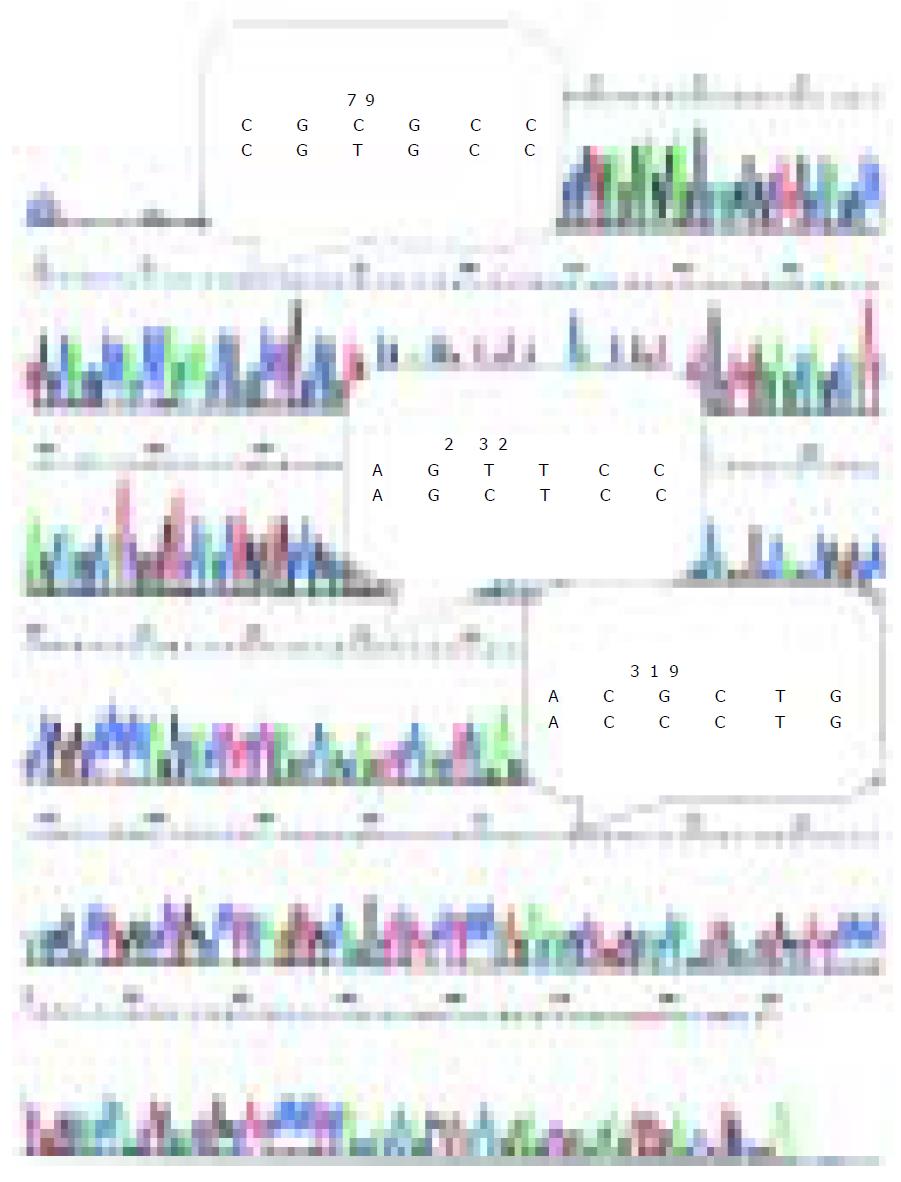
DcR3 gene amplification was proposed to be the mechanism for DcR3 overexpression to promote tumor survival in lung and colon cancer. To determine whether DcR3 overexpression was due to gene amplification, quantitative PCR was carried out on genomic DNA from all the patients, with the relative number DcR3 gene copies determined by normalization of the beta-globin gene. Unexpectedly, the amplification was 2-fold higher only in 7 of 48 tumors patients than that in 29 of 48 tumors patients with positive DcR3 mRNA expression. Among the 29 HCC cases with postive DcR3 mRNA expression, the amplification was 2-fold higher only in six cases. From the result of sequence analysis, we found accidentally that there were three bases different from the DcR3 mRNA sequence in GenBank (Figure 2).
In situ TUNEL was used to identify the apoptosis cells in 29 tissue specimens of HCC. The apoptosis index was calculated by counting the positive cells in five visual fields under light microscope at 400-fold magnification. DcR3 mRNA positive expression was detected in 19 out of 29 HCC cases. The apoptosis index of the 29 HCC cases ranged from 0.021 to 0.362. The apoptosis index of the 19 patients with positive DcR3 mRNA expression was significantly lower than that (0.067±0.04) of the 10 patients with negative DcR3 mRNA expression ( 0.209±0.12) (P<0.01).
Hepatoma cells of 19 HCC cases were studied. After treatment with FasL, the cell survival rate (CSR) of 10 HCC cases with DcR3 mRNA expression (83.7%±0.18) was significantly higher than that of another eight HCC cases without DcR3 expression (62.2%±0.16) (P<0.01).
From 0.25- to 4-fold drug concentrations, the hepatoma cell death induced by ADM, MMC, and GEMZAR had a positive relationship with the drug concentration (Table 2, P<0.01). But the effect of three drugs between hepatoma cells with positive and negative DcR3 mRNA expression had no significant difference (Table 3, P>0.05).
| ADM | p | MMC | p | GENZAR | p | |
| 4× | 51±0.2 | 44.6±0.19 | 52.2±0.17 | |||
| 1× | 72.7±0.18 | <0.01 | 64.8±0.16 | <0.01 | 66.7±0.17 | <0.01 |
| 0.25× | 82.6±0.17 | 81.8±0.16 | 77.9±0.18 |
| Survival rate of hepatoma cells | ||||||
| 4-fold | p | 1-fold | p | 0.25-fold | p | |
| ADM | ||||||
| DcR3 negative | 41.8±0.18 | >0.05 | 65.9±0.17 | >0.05 | 74.5±0.19 | >0.05 |
| DcR3 positive | 58.4±0.19 | 78.1±0.17 | 89.1±0.12 | |||
| MMC | ||||||
| DcR3 negative | 36.2±0.07 | >0.05 | 58.6±0.12 | >0.05 | 77.9±0.18 | >0.05 |
| DcR3 positive | 51.3±0.23 | 69.7±0.18 | 85±0.15 | |||
| GENZAR | ||||||
| DcR3 negative | 45.7±0.16 | 0.146 | 62.8±0.17 | 0.396 | 73.1±0.18 | 0.332 |
| DcR3 positive | 57.4±0.16 | 69.8±0.17 | 81.8±0.19 | |||
DcR3 gene is mapped on chromosome 20q13.3, a region known to be associated with gene amplification and rearrangement in human cancer[12,13]. Numerous reports have shown genomic amplifications of 20q13 in breast[14], gastric[15], colon, and lung[12] tumors and neuroblastomas[16]. Pitti et al[1] found that the DcR3 gene is amplified in approximately half of human lung tumors and human colon adenocarcinomas and that DcR3 receptor overexpression might also occur, conferring growth advantage on tumor cells by blocking FasL-induced cell death. In this paper, we demonstrated DcR3 mRNA and protein overexpression in HCC. From the result our data, we found that there were only 20.7% (6/29) genomic amplifications in HCC with positive DcR3 mRNA expression, suggesting that overexpression of DcR3 gene occurs without genomic amplification. Bai et al[2] examined DcR3 genomic DNA, mRNA, and protein levels in a series of human gastrointestinal tract tumors and found that DcR3 gene is overexpressed in a substantial number of tumors in which gene amplification could not be detected by fluorescence in situ hybridization or quantitative genomic PCR. The identification of tumors with high-level DcR3 overexpression but no significant gene amplification raises the possibility that DcR3 protein overexpression might be an early event in oncogenesis, possibly driving selection of gene amplification. The cause of nongenomic DcR3 overexpression remains unclear. From the result of sequence analysis, we found accidentally that there were three bases different from the DcR3 mRNA sequence in GenBank. The base difference might represent a point mutation or a gene polymorphism in HCC. The meaning and mechanism of the base differences in HCC remain unclear and still need further work.
Since DcR3 is a soluble protein, it can be easily detected in serum with DcR3 antibody. The protein overexpression is likely to be the eventual choice of diagnostic method for DcR3. We found a high DcR3 protein level in serum of the HCC patients by ELISA with anti-DcR3 antibody. Although the diagnostic value of DcR3 protein still needs an overall evaluation, at least it can be used as an auxiliary diagnostic method in combination with other methods for HCC.
Fas-induced apoptosis plays a role in the hepatic pathology of Wilson’s disease, in which Fas and FasL are induced in hepatocytes overloaded with copper[17,18]. Bai et al[2,19-21] used an model of copper-induced hepatocellular injury to test whether expression of DcR3 could protect cells from apoptosis and found that DcR3 might indeed plays a physiologically important role in the regulation of apoptosis. In this study, we found that DcR3 mRNA expression in HCC showed a significant relationship with the apoptosis index. Patients with a positive DcR3 mRNA expression had a low apoptosis index compared to those with a negative expression. The difference in apoptosis index between positive and negative DcR3 mRNA expression groups showed a statistic significance. suggesting that DcR3 protein on apoptosis shows its therapeutic value of down-regulating DcR3 overexpression by genetic or immunotherapeutic means.
Science Editor Wang XL Language Editor Elsevier HK
| 1. | Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396:699-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 588] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 2. | Bai C, Connolly B, Metzker ML, Hilliard CA, Liu X, Sandig V, Soderman A, Galloway SM, Liu Q, Austin CP. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci USA. 2000;97:1230-1235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 190] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 3. | Roth W, Isenmann S, Nakamura M, Platten M, Wick W, Kleihues P, Bähr M, Ohgaki H, Ashkenazi A, Weller M. Soluble decoy receptor 3 is expressed by malignant gliomas and suppresses CD95 ligand-induced apoptosis and chemotaxis. Cancer Res. 2001;61:2759-2765. [PubMed] |
| 4. | Yu KY, Kwon B, Ni J, Zhai Y, Ebner R, Kwon BS. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J Biol Chem. 1999;274:13733-13736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 295] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 5. | Ohshima K, Haraoka S, Sugihara M, Suzumiya J, Kawasaki C, Kanda M, Kikuchi M. Amplification and expression of a decoy receptor for fas ligand (DcR3) in virus (EBV or HTLV-I) associated lymphomas. Cancer Lett. 2000;160:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Otsuki T, Tomokuni A, Sakaguchi H, Aikoh T, Matsuki T, Isozaki Y, Hyodoh F, Ueki H, Kusaka M, Kita S. Over-expression of the decoy receptor 3 (DcR3) gene in peripheral blood mononuclear cells (PBMC) derived from silicosis patients. Clin Exp Immunol. 2000;119:323-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Ibrahim SM, Ringel J, Schmidt C, Ringel B, Müller P, Koczan D, Thiesen HJ, Löhr M. Pancreatic adenocarcinoma cell lines show variable susceptibility to TRAIL-mediated cell death. Pancreas. 2001;23:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Shen HW, Wu YL, Peng SY. [Overexpression and genomic amplification of decoy receptor 3 in hepatocellular carcinoma and significance thereof]. Zhonghua YiXue ZaZhi. 2003;83:744-747. [PubMed] |
| 9. | Wu Y, Han B, Sheng H, Lin M, Moore PA, Zhang J, Wu J. Clinical significance of detecting elevated serum DcR3/TR6/M68 in malignant tumor patients. Int J Cancer. 2003;105:724-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Tsuji S, Hosotani R, Yonehara S, Masui T, Tulachan SS, Nakajima S, Kobayashi H, Koizumi M, Toyoda E, Ito D. Endogenous decoy receptor 3 blocks the growth inhibition signals mediated by Fas ligand in human pancreatic adenocarcinoma. Int J Cancer. 2003;106:17-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Chen J, Zhang L, Kim S. Quantification and detection of DcR3, a decoy receptor in TNFR family. J Immunol Methods. 2004;285:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 12. | Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Identification of amplified DNA sequences in breast cancer and their organization within homogeneously staining regions. Genes Chromosomes Cancer. 1995;14:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, Nakamura Y, Inazawa J. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer. 1999;24:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 14. | Schwendel A, Richard F, Langreck H, Kaufmann O, Lage H, Winzer KJ, Petersen I, Dietel M. Chromosome alterations in breast carcinomas: frequent involvement of DNA losses including chromosomes 4q and 21q. Br J Cancer. 1998;78:806-811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Sakakura C, Mori T, Sakabe T, Ariyama Y, Shinomiya T, Date K, Hagiwara A, Yamaguchi T, Takahashi T, Nakamura Y. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;24:299-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 16. | Altura RA, Maris JM, Li H, Boyett JM, Brodeur GM, Look AT. Novel regions of chromosomal loss in familial neuroblastoma by comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:176-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Kondo T, Suda T, Fukuyama H, Adachi M, Nagata S. Essential roles of the Fas ligand in the development of hepatitis. Nat Med. 1997;3:409-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 393] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 18. | Strand S, Hofmann WJ, Grambihler A, Hug H, Volkmann M, Otto G, Wesch H, Mariani SM, Hack V, Stremmel W. Hepatic failure and liver cell damage in acute Wilson's disease involve CD95 (APO-1/Fas) mediated apoptosis. Nat Med. 1998;4:588-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 161] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Wu Y, Han B, Luo H, Roduit R, Salcedo TW, Moore PA, Zhang J, Wu J. DcR3/TR6 effectively prevents islet primary nonfunction after transplantation. Diabetes. 2003;52:2279-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Shi G, Wu Y, Zhang J, Wu J. Death decoy receptor TR6/DcR3 inhibits T cell chemotaxis in vitro and in vivo. J Immunol. 2003;171:3407-3414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Yang CR, Hsieh SL, Teng CM, Ho FM, Su WL, Lin WW. Soluble decoy receptor 3 induces angiogenesis by neutralization of TL1A, a cytokine belonging to tumor necrosis factor superfamily and exhibiting angiostatic action. Cancer Res. 2004;64:1122-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |