Published online Sep 28, 2005. doi: 10.3748/wjg.v11.i36.5651
Revised: February 13, 2005
Accepted: February 18, 2005
Published online: September 28, 2005
AIM: To study the effect of SNC19/ST14 gene overexpression on invasion in vitro of colorectal cancer cells.
METHODS: The adhesion of SNC19/ST14 gene-transfected cells to ECM was measured by MTT assay. The cell movement was evaluated by wound healing assay. Cell invasion and migration were determined by invasion assay in vitro.
RESULTS: SNC19/ST14 gene overexpression could enhance invasion of colorectal cancer cells in vitro significantly and influence early cell adherence to ECM, but could not change cell movement significantly.
CONCLUSION: SNC19/ST14 gene overexpression increases the local invasion of colorectal cancer cells in vitro.
- Citation: Ding KF, Sun LF, Ge WT, Hu HG, Zhang SZ, Zheng S. Effect of SNC19/ST14 gene overexpression on invasion of colorectal cancer cells. World J Gastroenterol 2005; 11(36): 5651-5654
- URL: https://www.wjgnet.com/1007-9327/full/v11/i36/5651.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i36.5651
SNC19/ST14 is recently found to be a new member of the serine protease family[1]. SNC19 gene has been isolated from a subtractive cDNA library of colorectal cancer by Cancer Institute of Zhejiang University, and is uploaded in the GenBank in 1995[2-4], and designated as ST14 (suppression of tumorigenicity 14) by Gene Nomenclature Committee in 1998[5]. From then on, homologous products of SNC19/ST14 have been found in many types of tissue cells, cell lines, and milk. SNC19/ST14 protein is designated as ST14/MT- SP1/matriptase/TADG-15, EC 3.4.21. SNC19/ST14 could convert hepatocyte growth factor (HGF)/scattering factor to its active form, and activate c-Met tyrosine phosphorylation[5]. Further, protease-activated receptor 2 (PAR2) and single-chain urokinase plasminogen activator (sc-uPA) have been identified as substrates of
SNC19/ST14/MT-SP1/matriptase/TADG-15[6].
The uPA and HGF/SF are closely correlated with ECM degradation, growth, movement, and metastasis of cancer cells[7]. The SNC19/ST14 protein could also directly degrade ECM[8,9]. Studies suggest that SNC19/ST14 is highly expressed in prostate, breast, and cervical cancer tissue[10-12]. The results suggest that SNC19/ST14 and its human ortholog may play an important role in cell migration, cancer invasion, and metastasis. To know the detailed information on how the SNC19/ST14 takes part in tumor invasion and metastasis, we compared the change of adhesion, movement, migration, and invasion of the stable ST14-transfected colorectal cancer cell clones to vector alone-transfected clones.
RKO, a colorectal cancer cell line, SNC19/ST14-transfected (RKO-ST14-1) and vector alone-transfected colorectal cancer cell clones (RKO-pSecTag) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (Gibcol Inc.), penicillin (50 U/mL), streptomycin (50 g/mL), glucose (4 500 mg/L), L-glutamine (2 mmol/L) and Zeocin™ (600 mg/mL), then incubated at 37°C in a humidified 95% air, 50 mL/L CO 2. Culture medium was changed on alternate days to avoid nutrient depletion.
Polystyrene 96-well plates (Costar) were coated with 50 μL/well of 8 μg/mL ECM (Sigma, USA), and left uncovered in a laminar flow hood overnight to allow evaporation. The plates were then rinsed with PBS and used for the attachment assays. Cells were washed thrice with PBS, trypsinized and seeded into 200 µL cells at a density of 2×105/mL on ECM. After 1, 2, 6, and 12 h of incubation at 37°C, the wells were gently rinsed thrice with PBS to remove unattached cells. The remaining cells in 96-well plates were reacted with MTT for 4 h at 37°C, then solved with DMSO. The absorbance of each well was measured at 570 nm. Results were expressed as the percentage of total cells assuming that the adhesion of cells in control was 100%. The percentage of adhesion was determined using the formula: (A570 nm after being rinsed with PBS/A570 nm no rinse)×100%. The experiments were performed in triplicate.
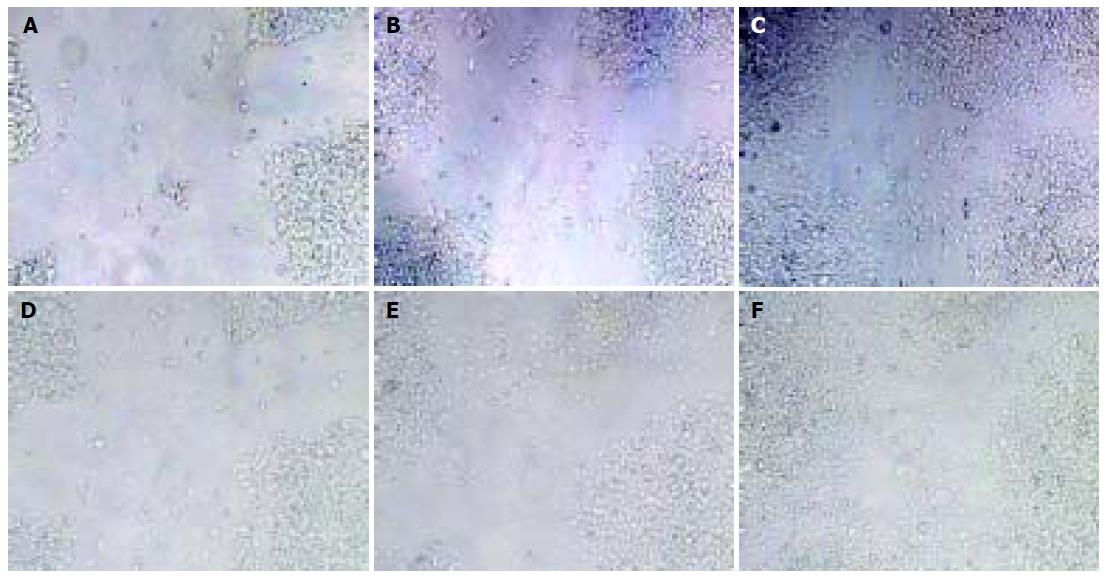
Confluent monoplayers were serum starved for 24 h and washed with PBS, and wounds were made with a pipette tip. After cell debris were removed, the cultures were incubated in DMEM alone or containing fetal calf serum. After 24 and 48 h, healing was evaluated under a phase contrast microscope and photographed at 200×magnification under a Zeiss microscope (Germany). The movement distance of the wounded cells was measured by ImageJ software.
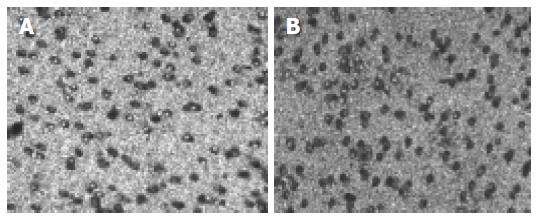
Invasion assays were carried out, following the manufacturer’s instructions of cell invasion assay kit (Chemicon International Inc., catalog: ECM550). For the invasion assay, we used a modified Boyden chamber. The chamber has two compartments divided by a polycarbonate filter (8 μm pore size) coated by a reconstituted basement membrane (ECMatrix™ solution). RKO, pSecTag vector controls (RKO- pSecTag), ST14-transfected cell clones (RKO-ST14-1) were grown in DMEM and harvested in PBS. After centrifugation the cell pellets were suspended in serum-free DMEM and 1.5×105 cells were placed into the upper chamber and 500 μL DMEM containing 10% fetal calf serum was added into the bottom chamber. Cells were incubated for 48 h at 37°C in 50 mL/L CO2. The non- invading cells and the ECMatrix gel were gently removed from the upper chamber with cotton-tipper swabs, and the filters were stained in the staining solution for 20 min and rinsed several times in water and air dried. Three filters were used for each cell type. The number of invading cells was counted in 10 random high-powered (HP) fields per filter under a Zeiss microscope.
The data were expressed as mean±SD and compared by ANOVA and Student’s t-test. P < 0.05 was considered statistically significant.
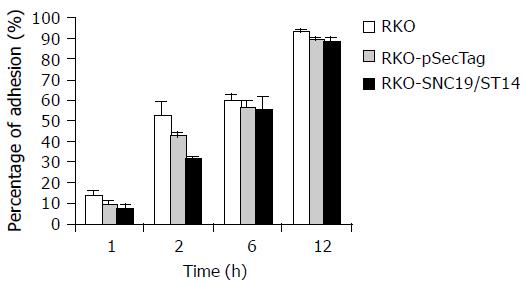
After 1 and 2 h, the percentage of ST14 transfectant (RKO-ST14-1) adhesion to ECM was about 5.1% and 32.3%, respectively, and significantly decreased compared to the wild type (RKO) and vector- transfected control (RKO-pSecTag) (P < 0.05). But after 6 and 12 h, their difference in cell adhesion to ECM was not significant (P > 0.05, Figure 1, and Table 1).
There was no significant difference in movement distance between RKO-ST14-1 and vector-transfected cells (RKO- pSecTag) (P > 0.05), but the movement distance of RKO-ST14-1 cells had a tendency to increase (Figure 2 and Table 2).
| Cell | 24 h | 48 h | P |
| R-P-media | 0.56 ± 0.24 | 0.44 ± 0.08 | 0.22 |
| R-S-media | 0.73 ± 0.56 | 0.72 ± 0.45 | 0.15 |
| R-P-serum | 0.97 ± 0.07 | 1.01 ± 0.27 | 0.37 |
| R-S-serum | 1.15 ± 0.46 | 1.11 ± 0.6 | 0.7 |
The number of invasive cells per HP field of SNC19/ST14 transfected clones (RKO- ST14-1), vector-transfected cells (RKO-pSecTag) and wild type cells (RKO) were 64.5 ± 7.3, 44.9 ± 7.0, and 32.6 ± 4.1, respectively. RKO- ST14-1 cells show a significantly increased the cell invasion as compared to wild type cells (RKO) and vector-transfected control (RKO-pSecTag) (Figure 3 and Table 3), suggesting that the ST14 protein might promote invasion of colorectal cancer cells.
| Cell line | Cell number/HP | P |
| RKO | 32.6 ± 4.1 | 0.000018 |
| RKO-pSecTag | 44.9 ± 7.0 | 0.00009 |
| RKO-ST14-1 | 64.5 ± 7.3 |
Tumor progression and metastasis can be considered as a cascade of events and a continuous selection process, which includes ECM recognition, matrix degradation, migration, hematogenous dissemination and organ selection metastasis[13]. It is accepted that malignant cell-ECM interaction must be considered as the key feature of malignancy. Malignant cells must recognize and degradate ECM before invasion and metastasis. Therefore, it is important to study matrix degrading enzymes for understanding and controlling the metastasis. The serine protease is a member of these ECM-degrading enzymes family. SNC19/ST14 protein has been recently found to be a new member of type II transmembrane serine protease (TTSPs)[1].
In our experiments, the SNC19/ST14 protein could enhance cell invasion in vitro when it was overexpressed in colorectal cancer cells. The final results of cell adhesion, degradation, and movement were evaluated by ECM invasion assay in vitro. In this study, although SNC19/ST14 overexpression could decrease cell adhesion to ECM in a short time, after 6 h this change was not observed. Besides, SNC19/ST14 overexpression could not increase cell movement. Since SNC19/ST14 overexpression could not induce cell cycle, apoptosis and proliferation, we believe that SNC19/ST14 enhancing cancer cell invasion results from increased ECM degradation.
There are several reasons to explain why SNC19/ST14 overexpression could increase ECM degradation. Firstly, SNC19/ST14 protein has extracellular matrix-degrading activity[8,9]. Secondly, SNC19/ST14 protein can activate HGF and uPA in vitro[6]. HGF and uPA have been implicated in cancer invasion and metastasis due to their cellular motility, extracellular matrix degradation, and tumor vascularization[14,15]. As a protease, uPA is best known to convert zymogen plasminogen into active plasmin. Plasmin promotes degradation of diverse ECM substrates such as fibrin, fibronectin, and laminin[16], activates certain matrix metalloproteases (MMPs) to allow further degradation of ECM, especially interstitial and type IV collagen. Therefore, uPA can promote metastasis by degrading ECM and permitting local invasion. Besides, HGF increases the expression of uPA and its receptor (uPAR)[18], and HGF precursor (pro-HGF) can be activated by uPA[19].
In conclusion, SNC19/ST14 can directly degrade ECM and activate uPA and HGF/SF to enhance ECM degradation, which might be one of the reasons why SNC19/ST14 overexpression enhances local invasion and tumor metastasis.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
Co-first-authors: Ke-Feng Ding and Li-Feng Sun
Co-correspondent: Su-Zhan Zhang
| 1. | Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem. 2001;276:857-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 276] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 2. | Cao J, Cai X, Zheng L, Geng L, Shi Z, Pao CC, Zheng S. Characterization of colorectal-cancer-related cDNA clones obtained by subtractive hybridization screening. J Cancer Res Clin Oncol. 1997;123:447-451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Zheng S, Cai X, Cao J, Zheng L, Geng L, Zhang Y, Gu J, Shi Z. Screening and identification of down-regulated genes in colorectal carcinoma by subtractive hybridization: a method to identify putative tumor suppressor genes. Chin Med J (Engl). 1997;110:543-547. [PubMed] |
| 4. | Zhang Y, Cai X, Schlegelberger B, Zheng S. Assignment1 of human putative tumor suppressor genes ST13 (alias SNC6) and ST14 (alias SNC19) to human chromosome bands 22q13 and 11q24--& gt; q25 by in situ hybridization. Cytogenet Cell Genet. 1998;83:56-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275:36720-36725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 295] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 6. | Takeuchi T, Harris JL, Huang W, Yan KW, Coughlin SR, Craik CS. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem. 2000;275:26333-26342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 311] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 7. | Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin Cancer Res. 2004;10:202-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 151] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Shi YE, Torri J, Yieh L, Wellstein A, Lippman ME, Dickson RB. Identification and characterization of a novel matrix-degrading protease from hormone-dependent human breast cancer cells. Cancer Res. 1993;53:1409-1415. [PubMed] |
| 9. | Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol Chem. 1999;274:18231-18236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 160] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 10. | Oberst M, Anders J, Xie B, Singh B, Ossandon M, Johnson M, Dickson RB, Lin CY. Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol. 2001;158:1301-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 150] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 11. | Bhatt AS, Takeuchi T, Ylstra B, Ginzinger D, Albertson D, Shuman MA, Craik CS. Quantitation of membrane type serine protease 1 (MT-SP1) in transformed and normal cells. Biol Chem. 2003;384:257-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Santin AD, Cane' S, Bellone S, Bignotti E, Palmieri M, De Las Casas LE, Anfossi S, Roman JJ, O'Brien T, Pecorelli S. The novel serine protease tumor-associated differentially expressed gene-15 (matriptase/MT-SP1) is highly overexpressed in cervical carcinoma. Cancer. 2003;98:1898-1904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Timar J, Csuka O, Orosz Z, Jeney A, Kopper L. Molecular pathology of tumor metastasis. Pathol Oncol Res. 2002;8:204-219. [RCA] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Comoglio PM, Boccaccio C. Scatter factors and invasive growth. Semin Cancer Biol. 2001;11:153-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Andreasen PA, Egelund R, Petersen HH. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell Mol Life Sci. 2000;57:25-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 705] [Cited by in RCA: 699] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 16. | Andreasen PA, Kjøller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 17. | Carmeliet P, Moons L, Lijnen R, Baes M, Lemaître V, Tipping P, Drew A, Eeckhout Y, Shapiro S, Lupu F. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet. 1997;17:439-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 477] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 18. | Jeffers M, Rong S, Vande Woude GF. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Mol Cell Biol. 1996;16:1115-1125. [PubMed] |