Published online Sep 14, 2005. doi: 10.3748/wjg.v11.i34.5385
Revised: January 23, 2005
Accepted: January 26, 2005
Published online: September 14, 2005
AIM: To examine the sensitivity and accuracy of real-time polymerase chain reaction (PCR) for the quantification of hepatitis B virus (HBV) DNA in semen.
METHODS: Hepatitis B viral DNA was isolated from HBV carriers’ semen and sera using phenol extraction method and QIAamp DNA blood mini kit (Qiagen, Germany). HBV DNA was detected by conventional PCR and quantified by TaqMan technology-based real-time PCR (quantitative polymerase chain reaction (qPCR)). The detection threshold was 200 copies of HBV DNA for conventional PCR and 10 copies of HBV DNA for real time PCR per reaction.
RESULTS: Both methods of phenol extraction and QIAamp DNA blood mini kit were suitable for isolating HBV DNA from semen. The value of the detection thresholds was 500 copies of HBV DNA per mL in the semen. The viral loads were 7.5 × 107 and 1.67 × 107 copies of HBV DNA per mL in two HBV infected patients’ sera, while 2.14 × 105 and 3.02 × 105 copies of HBV DNA per mL in the semen.
CONCLUSION: Real-time PCR is a more sensitive and accurate method to detect and quantify HBV DNA in the semen.
- Citation: Qian WP, Tan YQ, Chen Y, Peng Y, Li Z, Lu GX, Lin MC, Kung HF, He ML, Shing LK. Rapid quantification of semen hepatitis B virus DNA by real-time polymerase chain reaction. World J Gastroenterol 2005; 11(34): 5385-5389
- URL: https://www.wjgnet.com/1007-9327/full/v11/i34/5385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i34.5385
Human hepatitis B virus (HBV) is the major epidemiological agent of acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma[1-3]. At present, around 10-15% of individuals (estimated 200 million people) are chronically infected with HBV in China, and HBV-associated hepatocellular carcinoma (HCC) has become the country’s second most lethal disease.
One of the major HBV transmission pathways is parenteral[4,5]. HBV is therefore in most cases transmitted to individuals at birth or in the postnatal period by infected mothers, less commonly through close contact with infected fathers, siblings, and relatives during early childhood[2]. In developed countries, sexual transmission plays a major role in infecting individuals[3]. Mother-to-baby vertical transmission gives a large number of HBV carriers in China and other eastern Asian countries. It has also been reported that fetuses become infected in the uterus by their fathers as a result of transmission through sexual contact, although the mothers are negative for any HBV marker[6]. Therefore, the viral load in the semen or vaginal secretions is a very important parameter for safe sex and human reproduction.
Few attempts have been made to monitor HBV viral load in semen or vaginal secretions. Many people, especially in conservative Asian cultures, are reluctant to disclose information on their sexual habits, and unwilling to provide semen or vaginal secretions for epidemiological studies. When semen or vaginal secretions become available for study, they are in far smaller volumes than typical blood samples, and their viral titers are much lower than those of blood samples. These factors have hindered the progress of virological studies of reproduction-related body fluids. Although several methods have been developed for the detection of HBV DNA in semen[7-10], and HBV is routinely monitored when semen is screened for artificial insemination[11-13], the quantitative data produced have been disappointingly small. Southern blot has been used to estimate HBV viral load in semen in a study conducted 15 years ago[7], but no follow-up was made, probably because the work routinely monitored when semen is screened for artificial insemination[11-13], the quantitative data produced have been disappointingly small. Southern blot has been used to estimate HBV viral load in semen in a study conducted 15 years ago[7], but no follow-up was made, probably because the work involved would have been tedious, time-consuming, and possibly of limited accuracy. At present, there are still insufficient virological data to enable the risk factors for HBV infection through semen to be evaluated.
The sensitive quantitative real-time PCR (qPCR) provides an opportunity to investigate the viral load in the semen of HBV patients or carriers seeking assisted reproduction. The qPCR technology has greatly improved the precision of DNA quantification[14-16]. In this paper, we compared two different methods for the preparation of HBV DNA from HBV carriers’ semen, and presented a TaqMan technology-based assay to quantify HBV DNA in semen. Our assay is highly sensitive, and theoretically suitable for quantifying most HBV genotypes.
The study was conducted in patients who were seeking assisted reproduction in Luofu Hospital, Shenzhen, in 2003 and 2004. In accordance with the standard protocols, all patients who received assisted reproduction were systematically screened for serum hepatitis B virus surface antigen (HBsAg), hepatitis B surface antibody (HBsAb), hepatitis B e antigen (HBeAg), and hepatitis B e antibody (HBeAb) with commercially-available enzyme immunoassays (Abbott Laboratories, Chicago, IL, USA). The recommended viral markers including human immunodeficiency virus antibody (HIV Ab), and hepatitis C antibody (anti-HCV) were also detected. Hepatitis B core antibody (HBcAb) was also systematically tested by RIA (Corab; Abbott). Semen HBV DNA was detected by PCR. All patients were tested with liver function tests (including serum alanine aminotransferase, serum albumin and bilirubin) before assisted reproductions were carried out. Five patients were selected for this study. Two patients were positive for HBsAg, HBeAg and anti-HBc, and their serum ALT levels were normal over 6 mo. Two patients presented HBsAb only. One patient with all HBV markers negative (s, e, and c antibody negative; s and e antigene negative) was selected as a control (Table 1). HCV or HIV was not detected by RT-PCR in these patients’ sera. Patients were asked not to ejaculate for five days before semen samples were collected. Informed consent was obtained from all patients. Study procedures were approved by the Ethics Committee of Luofu Hospital and the Chinese University of Hong Kong. Each ejaculation was liquefied for at least 30 min at 37°C. The sperm concentration and percentage of motile spermatozoa were evaluated by Makler chamber, under a phase-contrast microscope. The serum and semen samples were stored at -20°C until use.
The spermatozoa in the semen were removed by centrifugation at 3 000 g at 4°C for 10 min. Different amounts of each seminal plasma (10, 50, and 200 μL) were used to isolate viral DNA by two methods. Viral DNA was isolated with QIAamp Blood Mini Kit (Qiagen, Germany) following manufacturer’s instructions. In the case of a small amount of semen, up to 200 μL of PBS was supplemented to avoid loss of viral DNA. In the phenol extraction experiments, an equal volume of saturated phenol (Sigma, St. Louis, USA) was added to the samples, and vortexed for 1 min. After centrifugation at 13 000 g for 10 min, upper layer was transferred to a fresh tube and repeated the extraction with phenol-chloroform (1:1). Then 0.1 volume of 3 mol/L potassium acetate (pH 5.2), 1 μL glycogen (1 μg/μL, invitrogen), and 2.5 volume of absolute alcohol were added to the extract, and frozen at -80°C for 20 min. The viral DNA was then precipitated by centrifugation at 13 000 g for 10 min, and finally resuspended in 20 μL TE buffer. As a control, 200 μL of each serum sample was used to isolate HBV DNA.
A plasmid containing Chinese HBV genome (pHBV-adr), kindly donated by Professor Yuan Wang[17], was isolated by CsCl purification. The DNA concentration was measured by A260 and verified by agarose gel electrophoresis. The copy number was determined by its molecule weight.
Previously, we used commercial detection kits to quantify HBV DNA in the sera[17]. To develop cheaper reagents and cover more HBV genotypes, we designed a set of primers and probes for this study. Various HBV genotypes undergo rapid mutagenesis because their reverse-transcriptase (RT) lacks proof-reading functions. To obtain specific PCR primers, we aligned over 150 sequences representing all the HBV genotypes, and designed a pair of primers for amplification of the S gene (conventional PCR) and core gene (quantitative PCR). Sequence alignments were carried out with the default settings using the BLAST algorithm (http://www.ncbi.nlm.nih.gov). Primers were chosen to facilitate amplification of most HBV genotypes. To achieve this goal, degenerated primers were designed. The primer pairs 5’-AGACTCGTGGTGGACTTCTCTC-3’ (forward) and 5’-AAGCCA(A/T/C/G)ACA(A/G)TGGGGGAAAGC-3’ (reverse) were used for amplification of the S gene in conventional PCR experiments while primers 5’-CCTGGGTGGGAAGTAATTTGG-3’ (forward), 5’-TTTTA(A/G)GCCCATATTAACATTGACAT-3’ (reverse), and TaqMan probe 5’-FAM-AGACCCA-GCATCCAGGGAATTAGTAGTCAGC-TAMRA-3’ were used to amplify core gene fragments in real-time PCR experiments. These primers could target different regions, thereby providing an important cross-check for the results.
Conventional PCR was used to detect HBV DNA in sera and semen. The PCR reactions were set in 20 µL volume containing 100 mmol/L Tris-HCl (pH 8.0), 2.5 mmol/L MgCl2, 1 unit of Taq DNA polymerase (Premega, Wisconsin, USA), 200 µmol/L each of the deoxynucleotide, triphosphates (dNTP), 1 µmol/L each of primers, and 1 µL viral DNA as template in each reaction. Thermal cycling included an initiation step at 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, at 60°C for 30 s, and at 72°C for 60 s. Products were visualized on agarose gels after electrophoresis.
Each 20-µL reaction contained 2 µL of sample extracts (template), 250 nmol/L of the probe, 900 nmol/L of each PCR primer, and 10 µL Master Mix (Roche, New Jersey, USA). PCR was performed using the ABI 7900HT sequence detection system. PCR cycling program consisted of an initial background eliminating step at 50°C for 5 min and a HotStart activation step at 95°C for 10 min, followed by 40 amplification cycles at 95°C for 15 s, and at 61°C for 40 s. The PCR products were further confirmed by electrophoresis loaded in 2% agarose gels. The experiments were repeated twice.
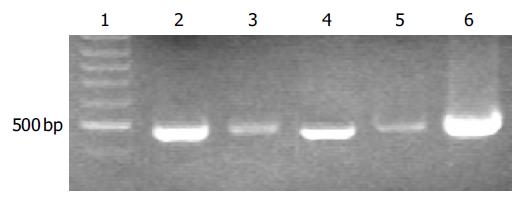
To detect hepatitis B viral DNA in the semen, HBV DNA was isloated from 200 µL of semen using QIAamp blood mini kit. Viral DNA was also isolated from the sera to serve as a control. Conventional PCR was carried out to amplify a 480-bp of DNA fragment. A specific fragment was amplified from viral DNA templates isolated from both sera and semen of HBV carriers (Patients 1 and 2), indicating that hepatitis B viruses existed in the semen of HBV carriers (Figure 1). The bands amplified from serum DNA (lanes 2 and 4) were much stronger than those amplified from semen DNA (lanes 3 and 5), indicating that the viral titers were much lower in semen than in sera. No DNA fragment was amplified from the samples (sera and semen) from the patient with HBsAb only or without any HBV markers (Patients 3-5, data not shown).
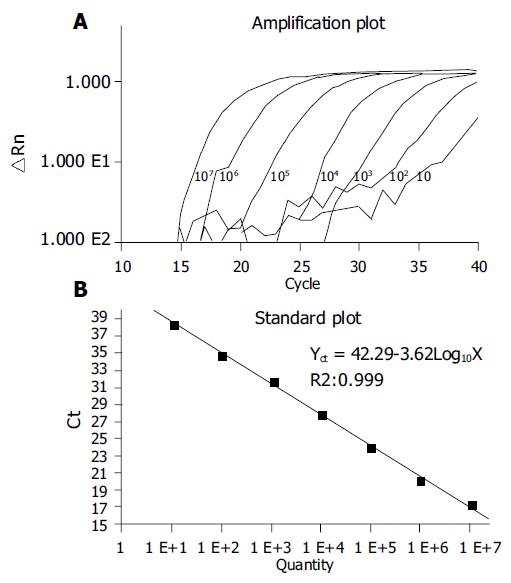
To test the performance of our primers and probes in real-time PCR, we used 10, 102, 103, 104, 105, 106, and 107 copies of HBV DNA[17]. Our results showed that amplification performance was good in all reactions (Figure 2A). A linear relationship was observed between 10 and 107 copies of HBV templates (Figure 2B). The viral titers in the sera and semen of patients were further quantified by real-time PCR. The consistent values of the qPCR were obtained by three independent qPCR (Table 2). We detected 7.56 × 107 copies of HBV per mL in the sera, but only 2.14 × 105 copies per mL in the semen of patient 1. Similar results were obtained from patient 2. We observed that the ratio of viral load in semen to sera was 0.3% in patient 1 and 1.8% in patient 2, respectively. The viral DNA isolations with different methods did not result in a statistically-significant difference (Student’s t-test). As we expected, no viral DNA was detected in the samples from non-infected patients (patient 5) or from the patient with positive HBsAb only (patients 3 and 4).
| Sample1 | Method of HBV DNA Isolation | HBV DNA (copies/mL)2 | Mean | SD3 | ||
| 1 | 2 | 3 | ||||
| Sera-1 | Qiagen kit | 6.73 × 106 | 7.14 × 106 | 8.8 × 106 | 7.56 × 107 | 1.11 × 107 |
| Semen-1 | Qiagen kit | 2.18 × 105 | 2.34 × 105 | 1.69 × 105 | 2.14 × 105 | 4.31 × 104 |
| Semen-1 | Phenol | 1.82 × 105 | 2.31 × 105 | 1.98 × 105 | 2.04 × 105 | 2.52 × 104 |
| Sera-2 | Qiagen kit | 1.82 × 107 | 1.73 × 107 | 1.47 × 107 | 1.67 × 107 | 1.82 × 106 |
| Semen-2 | Qiagen kit | 2.81 × 105 | 2.59 × 105 | 3.67 × 105 | 3.02 × 105 | 5.71 × 104 |
| Semen-2 | Phenol | 3.58 × 105 | 2.62 × 105 | 2.41 × 105 | 2.87 × 105 | 5.22 × 105 |
To validate whether our methods could apply to low-volume semen samples, we isolated HBV DNA from 10, 50 and 200 μL of semen and quantified HBV DNA copies by real-time PCR. Again, all experiments produced consistent results (Table 3), indicating that our methods could indeed be used to quantify HBV DNA with low-volume semen samples.
| Sample | Method of HBVDNA isolation | 10 μL | 50 μL | 200 μL |
| Semen-1 | Qiagen kit | 2.37 × 105 | 2.26 × 105 | 2.43 × 105 |
| Convention | 2.25 × 105 | 3.04 × 105 | 1.80 × 105 | |
| Semen-2 | Qiagen kit | 2.76 × 105 | 2.92 × 105 | 2.87 × 105 |
| Convention | 2.38 × 105 | 2.56 × 105 | 2.74 × 105 |
In this study, we found that TaqMan technology-based real-time PCR assay could be used to quantify HBV DNA in semen. As this method is highly sensitive and reproducible, and has a wide dynamic linear range, it can be used to monitor HBV viral load in small samples of body fluids. The detection threshold of this assay is 10 copies of HBV DNA per reaction.
Traditionally, HBV in sera is monitored by antigen/ antibody test, Dot blot, Southern blot, branched DNA assay, or hybrid capture test[2,18,19]. The minimal template requirement for these assays on blood is at least 7 × 105 copies of HBV DNA per millilter. The threshold of Roche Cobas RT-PCR commercial kit is 400 copies and that of New England DNA kit is around 100 copies. Due to sensitivity limitations, these requirements often give rise to problems in clinical settings. A large portion of HBV patients with undetectable viral antigens or viral DNA often relapse after the cessation of anti-HBV treatments[19], and for this reason real-time PCR has been recently introduced to monitor HBV viral load in sera[18,20]. In this study, we introduced real-time PCR method to quantify HBV DNA in semen of HBV carriers. Possible PCR inhibitors in the seminal extracts may cause false results. To exclude this possibility, we measured viral load from a sample extracted by different methods, as well as by the same method but with a different amount of samples. Our assay produced consistent reproducible results (Tables 2 and 3). These results indicated that this method is highly accurate and reproducible.
Theoretically, our quantification method may be used to quantify HBV DNA with multiple genotypes or mutants. Since HBV genome undergoes fast mutagenesis, the designed PCR primers may not match to a special HBV template or a mutated HBV template with reduced sensitivity. To make our assay applicable to most HBV genotypes, we aligned multiple HBV genome sequences containing most genotypes, and then designed primers targeting the most conserved core region. In addition, we defined the hot spot mutation sites in the HBV genome and designed degenerated PCR primers that could match the natural HBV templates or accumulated HBV mutations during anti-HBV treatment.
The use of TaqMan probe-based real-time PCR to quantify HBV DNA in body fluids has a number of advantages. Firstly, it is highly sensitive and accurate. As shown in Figure 2, as few as 10 copies of HBV template (corresponding to 500 copies of HBV DNA/mL semen) were sufficient to produce a marked amplification profile in the PCR reaction, and a wide linear range from 10 to 107 copies of HBV DNA was obtained. Real-time PCR can therefore be used to monitor HBV DNA in small samples of body fluid. Secondly, it has a low background. We noticed that there was no detectable HBV DNA either in the sera of non-HBV-carriers or in the semen of patients 3-5. When reaction and detection are carried out in the same reaction tube, the chance of contamination is greatly reduced. Thirdly, it could produce a very small number of false positive results. Positive results can only be obtained when PCR primers and molecular probes work in a small genomic element simultaneously. The chance of three specific primers targeting any other templates is very low. In our experiment, HBV DNA was not detected in the samples from non-HBV-infected patients. Fourthly, no PCR inhibitors were found in the semen, because consistent results were obtained from different DNA isolation methods. Finally, the method is quick and easy, and has a high throughput. A large number of samples can be simultaneously tested on a 96-well plate within 2-3 h. This is a particularly important consideration in clinical settings. There are about 200 million HBV carriers in China, and doctors need to know the HBV status of patients before medical treatments such as assisted reproduction, surgery, transfusion, and transplantation. These advantages make a popular way of quantifying HBV DNA in body fluids in the near future.
In summary, quantitative real-time PCR is a simple, sensitive, specific, and reproducible assay for the measurement of HBV viral load in semen. The viral load in body fluids, including semen and vaginal secretions, is an important parameter for the estimation of risk factor for HBV transmission through sexual contact.
Science Editor Wang XL and Li WZ Language Editor Elsevier HK
Co-first-authors: Wei-Ping Qian and Yue-Qiu Tan
| 1. | Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907-1917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3241] [Cited by in RCA: 3282] [Article Influence: 149.2] [Reference Citation Analysis (0)] |
| 2. | Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362:2089-2094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 590] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 3. | Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1728] [Cited by in RCA: 1712] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 4. | Spada E, Mele A, Ciccozzi M, Tosti ME, Bianco E, Szklo A, Ragni P, Gallo G, Balocchini E, Sangalli M. Changing epidemiology of parenterally transmitted viral hepatitis: results from the hepatitis surveillance system in Italy. Dig Liver Dis. 2001;33:778-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Judson FN. Epidemiology of sexually transmitted hepatitis B infections in heterosexuals: a review. Sex Transm Dis. 1981;8:336-343. [PubMed] |
| 6. | Wang S, Peng G, Li M, Xiao H, Jiang P, Zeng N, Wang Z. Identification of hepatitis B virus vertical transmission from father to fetus by direct sequencing. Southeast Asian J Trop Med Public Health. 2003;34:106-113. [PubMed] |
| 7. | Jenison SA, Lemon SM, Baker LN, Newbold JE. Quantitative analysis of hepatitis B virus DNA in saliva and semen of chronically infected homosexual men. J Infect Dis. 1987;156:299-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Davison F, Alexander GJ, Trowbridge R, Fagan EA, Williams R. Detection of hepatitis B virus DNA in spermatozoa, urine, saliva and leucocytes, of chronic HBsAg carriers. A lack of relationship with serum markers of replication. J Hepatol. 1987;4:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Karayiannis P, Novick DM, Lok AS, Fowler MJ, Monjardino J, Thomas HC. Hepatitis B virus DNA in saliva, urine, and seminal fluid of carriers of hepatitis B e antigen. Br Med J (Clin Res Ed). 1985;290:1853-1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 60] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Scott RM, Snitbhan R, Bancroft WH, Alter HJ, Tingpalapong M. Experimental transmission of hepatitis B virus by semen and saliva. J Infect Dis. 1980;142:67-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 116] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Eastlund T. Infectious disease transmission through cell, tissue, and organ transplantation: reducing the risk through donor selection. Cell Transplant. 1995;4:455-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Berry WR, Gottesfeld RL, Alter HJ, Vierling JM. Transmission of hepatitis B virus by artificial insemination. JAMA. 1987;257:1079-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Mascola L, Guinan ME. Screening to reduce transmission of sexually transmitted diseases in semen used for artificial insemination. N Engl J Med. 1986;314:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 34] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 14. | Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899-2903. [PubMed] |
| 15. | Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4152] [Cited by in RCA: 3922] [Article Influence: 135.2] [Reference Citation Analysis (1)] |
| 16. | Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1437] [Cited by in RCA: 1409] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 17. | He ML, Wu J, Chen Y, Lin MC, Lau GK, Kung HF. A new and sensitive method for the quantification of HBV cccDNA by real-time PCR. Biochem Biophys Res Commun. 2002;295:1102-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Xunrong L, Yan AW, Liang R, Lau GK. Hepatitis B virus (HBV) reactivation after cytotoxic or immunosuppressive therapy--pathogenesis and management. Rev Med Virol. 2001;11:287-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 78] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Lau GK, He ML, Fong DY, Bartholomeusz A, Au WY, Lie AK, Locarnini S, Liang R. Preemptive use of lamivudine reduces hepatitis B exacerbation after allogeneic hematopoietic cell transplantation. Hepatology. 2002;36:702-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 128] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Lewin SR, Ribeiro RM, Walters T, Lau GK, Bowden S, Locarnini S, Perelson AS. Analysis of hepatitis B viral load decline under potent therapy: complex decay profiles observed. Hepatology. 2001;34:1012-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 152] [Article Influence: 6.3] [Reference Citation Analysis (0)] |