Published online Sep 14, 2005. doi: 10.3748/wjg.v11.i34.5295
Revised: October 23, 2004
Accepted: October 26, 2004
Published online: September 14, 2005
AIM: To generate and characterize the synthetic transcriptional control units for transcriptional targeting of the liver, thereby compensating for the lack of specificity of currently available gene therapeutic vector systems.
METHODS: Synthetic transcriptional control unit constructs were generated and analyzed for transcriptional activities in different cell types by FACS quantification, semi-quantitative RT-PCR, and Western blotting.
RESULTS: A new bifunctionally-enhanced green fluorescent protein (EGFP)/neor fusion gene cassette was generated, and could flexibly be used both for transcript quantification and for selection of stable cell clones. Then, numerous synthetic transcriptional control units consisting of a minimal promoter linked to “naturally” derived composite enhancer elements from liver-specific expressed genes or binding sites of liver-specific transcription factors were inserted upstream of this reporter cassette. Following liposome-mediated transfection, EGFP reporter protein quantification by FACS analysis identified constructs encoding multimerized composite elements of the apolipoprotein B100 (ApoB) promoter or the ornithin transcarbamoylase (OTC) enhancer to exhibit maximum transcriptional activities in liver originating cell lines, but only background levels in non-liver originating cell lines. In contrast, constructs encoding only singular binding sites of liver-specific transcription factors, namely hepatocyte nuclear factor (HNF)1, HNF3, HNF4, HNF5, or CAAT/enhancer binding protein (C/EBP) only achieved background levels of EGFP expression. Finally, both semi-quantitative RT-PCR and Western blotting analysis of Hep3B cells demonstrated maximum transcriptional activities for a multimeric 4xApoB cassette construct, which fully complied with the data obtained by initial FACS analysis.
CONCLUSION: Synthetic transcriptional control unit constructs not only exhibit a superb degree of structural compactness, but also provide new means for liver-directed expression of therapeutic genes.
- Citation: Lemken ML, Wybranietz WA, Schmidt U, Graepler F, Armeanu S, Bitzer M, Lauer UM. Liver-directed gene expression employing synthetic transcriptional control units. World J Gastroenterol 2005; 11(34): 5295-5302
- URL: https://www.wjgnet.com/1007-9327/full/v11/i34/5295.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i34.5295
One of the major challenges in liver gene therapy is the achievement of hepato-specific therapeutic gene expression[1,2]. In general, transcriptional targeting is highly desirable for all in vivo gene therapy applications as it can prevent expression of the transgene in non-target cells, thus mimicking physiological regulation[3-5].
For the liver, numerous approaches to incorporate house-keeping cellular promoters or organ-specific regulatory sequences into retroviral, lentiviral or adenoviral vectors have been reported so far[6-8], with variable and repeatedly disappointing results. On one hand, the level of expression does not approach that of strong viral promoters such as human cytomegalovirus immediate early gene (hCMV IE) promoter. On the other hand, a functional tissue specificity only rarely has been demonstrated. As a consequence, the search for effective liver-restricted promoters has been extended to intensive screening of liver-expressed sequence tag (EST)-libraries still awaiting breakthrough[9,10].
For these reasons, promoter units providing sufficient transcriptional activity and concomitantly also tissue specificity still are required. Such promoter units can be built up synthetically on so-called minimal promoters, providing a basic/minimal transcriptional activity, which are combined with enhancer elements of either natural or synthetic origin being composed of suitable transcription factor binding sites (TFBS). This alternative approach should enable both a high degree of structural compactness and a high degree of liver specificity[11-13].
We here report the results of a comparative study addressing this issue. We employed a minimal core promoter derived from the Herpes simplex virus thymidine kinase promoter (HSV TK-37) which was combined in various arrangements with either “naturally” derived composite enhancer elements from liver-specific expressed house-keeping ally genes or binding sites for transcription factors guiding expression of liver-specific genes.
The enhancer elements of natural origin employed in this work were from the liver-specific human apolipoprotein B100 (ApoB) gene promoter which is located upstream at the region -109-55 of the transcription start site and exhibits binding sites for liver-specific transcription factors HNF1, HNF3, HNF5, CAAT/enhancer binding protein (C/EBP)[14-16] and from the liver-specific enhancer of the rat ornithine transcarbamoylase (OTC) gene promoter which is located 11 kbp upstream of the transcription start site and exhibits binding sites for liver-specific transcription factors HNF4 and C/EBP[17].
Sequences for single binding sites for liver -specific transcription factors were taken from the human albumin gene promoter (TFBS sequences for HNF1 and C/EBP[18]) and from the ApoB gene promoter (TFBS sequences for HNF3/4/5[14-16]).
All newly generated constructs were analyzed for transcriptional activity in different cell types in cell culture.
All synthetic transcriptional control unit constructs were based on a Herpes simplex virus thymidine kinase derived minimal core promoter (HSV TK-37), which only comprises the RNA initiation site and 37 upstream base pairs[19]. As a basic construct for the generation of new reporter plasmids, we used plasmid pTK-37CAT, in which the HSV TK-37 minimal core promoter drives expression of the chloramphenicol acetyltransferase (CAT) reporter gene[20].
Plasmid pEGFP-C1 (BD Biosciences Clontech, Heidelberg, Germany) facilitating the creation of protein fusions to the C-terminus of enhanced green fluorescent protein (EGFP), which constitutes a human codon-optimized cDNA of GFP[21,22] was used to generate a new EGFP/neor fusion reporter gene. For this purpose, the neomycin aminoglycoside phosphotransferase coding sequence of transposon 5 (i.e., neomycin resistance-neor) was cut out of plasmid pNEO (Pharmacia, Freiburg, Germany) and inserted in frame downstream of the EGFP coding sequence. This EGFP/ neor fusion gene was cut out and used for exchange with the CAT coding sequence in pTK-37CAT, thus a new reporter construct pGreeN (i.e., encoding the “GreeN(eo) fusion”) was yielded. Since expression of EGFP in pEGFP-C1 is driven by the strong, constitutive hCMV IE promoter, this vector was used also as a simple transfection marker for all cell lines of interest.
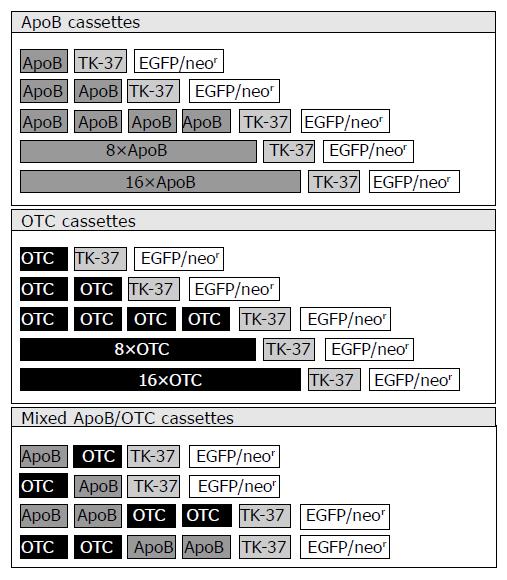
Since pGreeN was designed to exhibit a multiple cloning site (MCS) upstream of the HSV TK-37 minimal core promoter, insertion of composite enhancer elements from liver-specifically expressed genes or TFBS sequences via BamHI and BglII endonuclease restriction sites was facilitated. Insertion of complementary oligonucleotides encoding the binding sites of liver-specific transcription factors, namely hepatocyte nuclear factor 1 (HNF1; TFBS sequence originating from the human albumin gene promoter), hepatocyte nuclear factors 3/4/5 (HNF3/4/5; TFBS sequences all originating from the human apolipoprotein B100 gene promoter), C/EBP (TFBS sequence originating from the human albumin gene promoter) led to the generation of synthetic transcriptional control unit constructs p1 × HNF1-GreeN, p3 × HNF1-GreeN (exhibiting three consecutive HNF1 TFBS), p1 × HNF3-GreeN, p1xHNF4-GreeN,p1 × HNF5-GreeN,p1 × C/EBP-GreeN,respectively. Insertion of “natural” composite enhancer elements being important for liver-specific expression of the ApoB gene (60 bp; encoding TFBS for HNF3, HNF4, HNF5, and C/EBP) or the OTC gene (52 bp; encoding TFBS for HNF4 and C/EBP) also was inserted upstream of the HSV TK-37 minimal core promoter giving rise to synthetic transcriptional control unit constructs p1 × ApoB-GreeN and p1 × OTC-GreeN (Figure 1; first lines of upper and middle panel). Finally, plasmids encoding up to 16 monotype ApoB cassettes (Figure 1; upper panel), up to 16 monotype OTC cassettes (Figure 1; middle panel), and mixed ApoB/OTC cassettes (Figure 1; lower panel; exhibiting up to 4 mixed ApoB and OTC units) were constructed. All PCR-derived segments/constructs were sequenced thereby confirming their predicted composition (More detailed information about the construction process is available from the authors upon request).
Cell lines HepG2 (human hepatocellular carcinoma), Hep3B (human hepatocellular carcinoma), HeLa (human cervix carcinoma), HEK293 (human embryonic kidney) all were purchased from the European Collection of Cell Cultures (ECACC). Cell line HuH7 (human hepatocellular carcinoma) was generated originally and provided by Nakabayashi[23]. All cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37 in 50 mL/L CO2. No antibiotics were added.
Cells were plated at 3 × 105 cells/well into 6 -well trays (Falcon-Becton Dickinson Labware, Franklin Lakes, NJ, USA) and transfected 24 h later with 2 µg DNA complexed with either the multi-component lipid-based FuGENE 6™ transfection reagent (Roche Diagnostics, Mannheim, Germany), the cationic lipid reagent Lipofectamine™ 2000 (Invitrogen, Karlsruhe, Germany), the 1:1 (w/w) DOTMA/ DOPE liposome Lipofection™ reagent (Invitrogen), the polycationic DOSPER™ reagent (Boehringer, Mannheim, Germany), or the monocationic cholesterol DAC-30™ reagent (Eurogentec, Seraing, Belgium), according to the protocols of the manufacturers. To determine the transfection efficiencies, two separate transfections with samples were performed in triplicates, respectively.
Forty-eight hours after transfection, cells were washed with PBS, trypsinized and spun by centrifugation at 1200 r/min for 2 min. Cell pellets were resuspended in 500 µL PBS. The percentage of EGFP expressing cells was calculated by FACS analysis using the FL1-H profile employing a flow cytometer (FACS Calibur™, BD Biosystems, Heidelberg, Germany) with the CellQuest software according to the manufacturer’s instructions. To standardize the transfection efficiencies, all cell lines obtained underwent triple transfections with hCMV IE-driven control plasmid pEGFP-C1. The resulting pEGFP- C1-mediated mean EGFP positive cells were set to 100%; EGFP positive cells obtainedbytransfectionwithsynthetictranscriptionalcontrol unit constructs were set in relation to this EGFP standard level, thereby defining the ratio of relative EGFP expression. Data plotted represented the mean±SD of two separate transfections performed in triplicates, respectively.
Total RNA was extracted 48 h after transfection with the NucleoSpin RNA II kit (Macherey Nagel, Düren, Germany). Reverse transcription was performed using the SuperScript™ II RT kit (Invitrogen, Karlsruhe, Germany). Semi-quantitative PCR analysis was done using the Taq DNA polymerase kit (QIAGEN, Hilden, Germany). All protocols were performed according to the manufacturer’s instructions. The sequences of primers for semi-quantitative PCR are as follows. The 5’ primer for amplification of the EGFP gene is 5’-ATG GTG AGC AAG GGC GAG GAG CTG TTC-3’ and the 3’ primer is 5’-CTT GTA CAG CTC GTC CAT GCC GAG AGT-3’, giving rise to a specific band of 717 bp; the 5’ primer for amplification of the α-tubulin house-keeping gene (used as an internal standard) is 5’-ATG CCA AGC GTG CCT TTG TTC ACT GGT AC-3’ and the 3’ primer is 5’-GAA ATT CTG GGA GCA TGA CAT GCT GCA G -3’, giving rise to a specific band of 208 bp.
Forty-eight hours after transfection, cells were washed in PBS and trypsinized, proteins were extracted with 120 µL lysis buffer (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1% Nonidet P- 40, 200 mmol/L PMSF). Then 50 µL of cell lysates was loaded onto a 10% SDS polyacrylamide gel and separated by electrophoresis and transferred to a polyvinylidene difluoride membrane (Amersham Pharmacia Biotech, Freiburg, Germany) . After the non-specific binding sites were blocked with 5% non-fat milk in TBS-T (10 mmol/L Tris-HCl, 150 mmol/L NaCl with 0.05% Tween20) overnight at 4 , the membrane was incubated for 3 h with monoclonal mouse anti-α-tubulin antibody (Sigma, Saint Louis, USA) and monoclonal mouse anti-GFP antibody (Roche Applied Science, Indianapolis, USA). Proteins were visualized using horseradish peroxidase-conjugated anti- mouse antibody (BioRad, Hercules, USA) diluted in TBS for 45 min at room temperature followed by enhanced chemoluminescence (ECL; Amersham Pharmacia Biotech, Freiburg, Germany).
To facilitate both the quantification of transcriptional activities of newly designed synthetic transcriptional control cassettes and the selection of stable cell clones transfected with such constructs, a new bifunctional EGFP/neor fusion gene cassette was generated. Employing plasmid pEGFP-C1, a neomycin aminoglycoside phosphotransferase coding sequence, was linked in frame downstream of the EGFP coding sequence. Subsequently, this EGFP/neor fusion gene was cut out and used for exchange with the CAT coding sequence encoded in reporter plasmid pTK-37CAT[20], yielding the new reporter construct pGreeN, which also exhibits a multiple cloning site (MCS) upstream of the HSV TK-37 minimal core promoter.
New reporter construct pGreeN was used to generate synthetic transcriptional control unit constructs. For this purpose, either composite enhancer elements from liver-specifically expressed genes or binding site sequences of transcription factors guiding expression of liver-specific genes were inserted into the MCS-located upstream of the HSV TK-37 minimal core promoter.
Insertion of “natural” composite enhancer elements which are important for liver-specific expression of the human apolipoprotein B100 (ApoB) or the rat ornithin transcarbamoylase (OTC), generated synthetic transcriptional control unit constructs pApoB-GreeN and pOTC-GreeN. In addition, plasmids encoding up to 16 monotype ApoB cassettes, up to 16 monotype OTC cassettes, as well as mixed ApoB/OTC cassettes were constructed (Figure 1).
Insertion of complementary oligonucleotides encoding the binding sites of liver-specific transcription factors HNF1/3/4/5 or C/EBP led to the generation of synthetic transcriptional control unit constructs p1 × HNF1-GreeN, p3 × HNF1-GreeN (exhibiting three consecutive HNF1 TFBS), p1 × HNF3-GreeN, p1 × HNF4-GreeN, p1 × HNF5-GreeN, p1 × C/EBP-GreeN, respectively (constructs not depicted).
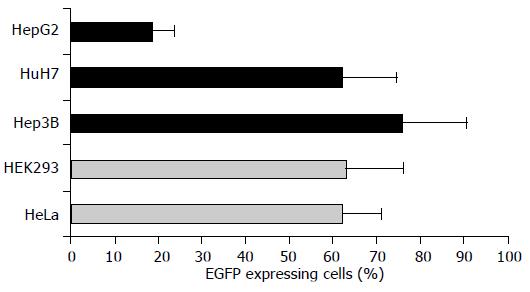
To evaluate the tissue-related transcriptional activities of pGreeN-based synthetic transcriptional control unit constructs, three human hepatoma cell lines (HepG2, Hep3B, HuH7) were chosen and compared with two carcinoma cell lines of cervix (HeLa) and human embryonic kidney (HEK293) origin, respectively. By mixing reporter plasmid pEGFP-C1 with five different commercially available lipid-based transfection reagents (FuGENE 6™, Lipofectamine 2000™, Lipofection™, DOSPER™, DAC- 30™) followed by subsequent FACS quantitation of EGFP expressing cells, we identified the multi-component lipid-based FuGENE 6 transfection reagent and yielded the best overall transfection efficiencies considering all five cell lines tested (data not shown). In particular, the transfection efficiencies obtained by FuGENE 6 were in the range of 60-75% for four of five cell lines tested (Figure 2; cell lines Hep3B, HuH7, HeLa, HEK293). Only HepG2 cells yielded a significant lower transfection efficiency of nearly 20% (Figure 2; first bar). However, none of the other transfection reagents tested was found to be able to achieve a roughly equivalent transfection efficiency in HepG2 cells (data not shown). Therefore, all successive transfection experiments were performed using FuGENE 6. Cell line-dependent differences in transfection efficiencies were normalized by setting EGFP expression levels obtained with hCMV IE-driven control plasmid pEGFP-C1 to 100% for each cell line tested. EGFP expression levels obtained by transfection with synthetic transcriptional control unit constructs were related to this cell line-dependent EGFP standard level.
Following FuGENE 6-mediated transfection of cell lines HepG2, Hep3B, HuH7, HeLa, HEK293, with synthetic transcriptional control unit constructs, percentages of EGFP expressing cells were determined in a simplified first round of testing by FACS analysis, constituting a first rough correlation for the individual transcriptional activities of each construct.
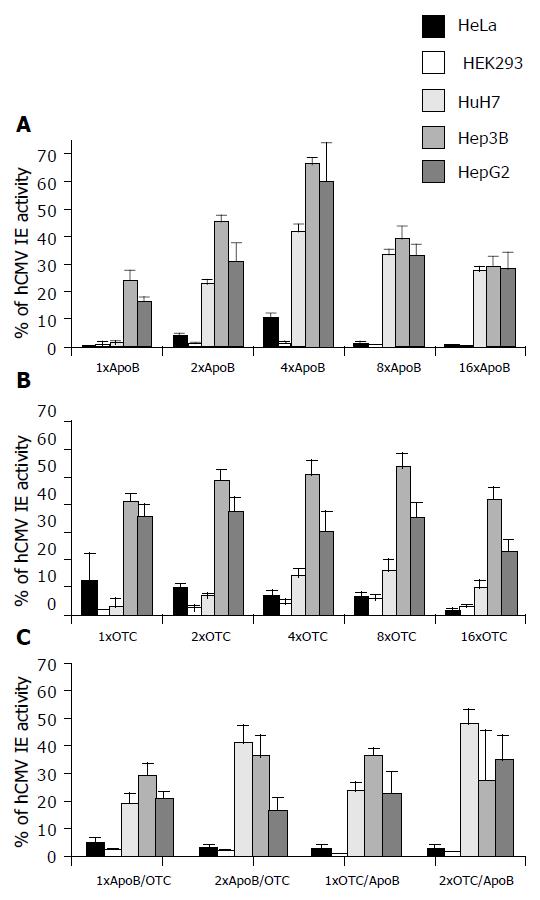
As a result, constructs comprising monotype ApoB cassettes (Figure 3; upper panel) exhibited high EGFP levels of over 60% (Figure 3; Hep3B and HepG2 cells transfected with 4 × ApoB cassette construct) only in human hepatoma derived cell lines. In contrast, EGFP expression of non-liver origin cell lines HeLa and HEK293 transfected with monotype ApoB cassette constructs only reached background levels below or around 10% of cell line-dependent EGFP standard levels (Figure 3; black and grey bars in upper panel).
Therefore, monotype ApoB cassette constructs (encoding TFBS for HNF3, HNF4, HNF5, and C/EBP) could provide a strong liver-directed gene expression. Interestingly, maximum hepatoma cell-based reporter gene expression was obtained with the 4 × ApoB cassette construct (Figure 3). In contrast, constructs exhibiting 8 or 16 multimeric ApoB cassettes only demonstrated EGFP expression levels between 30% and 40% or just below 30%, respectively, a phenomenon which could be referred to excess presentation of TFBS encoded by these highly multimeric ApoB cassette constructs.
Constructs comprising monotype OTC cassettes (Figure 3; middle panel) exhibited high EGFP-levels of over 50% (Figure 3; Hep3B cells transfected with 4 × or 8 × OTC cassette constructs) only in human hepatoma derived cell lines. EGFP expression of non-liver origin cell lines HeLa and HEK293 transfected with monotype OTC cassette constructs only reached background levels below or around 10% of cell line-dependent EGFP standard levels (Figure 3; black and grey bars in middle panel).
Therefore, monotype OTC cassette constructs (encoding TFBS for HNF4 and C/EBP) could also provide a strong liver-directed gene expression in Hep3B and HepG2 hepatoma cells. On the contrary, HuH7 cells transfected with monotype OTC cassette constructs exhibited only EGFP expression levels of up to 16% (Figure 3; middle panel transfected with 8 × OTC cassette construct). Again, all hepatoma cell lines (HepG2, Hep3B, HuH7) transfected with construct p16 × OTC-GreeN exhibiting 16 multimeric OTC cassettes demonstrated lower EGFP expression levels in comparison to transfections with constructs p4 × OTC-GreeN and p8 × OTC-GreeN (Figure 3; middle panel), a phenomenon which again could be referred to excess presentation of TFBS encoded by this highly multimeric (16 × ) OTC cassette construct.
Transfection experiments with non-liver origin cell lines HeLa and HEK293 employing mixed ApoB/OTC cassette constructs (Figure 3; lower panel; exhibiting up to 4 ApoB/OTC units) exhibited EGFP expression levels being unexceptionally below the 5% margin of cell line-dependent EGFP standard levels. In contrast, maximum EGFP expression levels between 30% and nearly 50% were obtained in hepatoma cell lines HepG2, Hep3B, and HuH7 (Figure 3; yellow, orange and brown bars in lower panel). Therefore, mixed ApoB/OTC cassette constructs (encoding TFBS for HNF3, HNF4, HNF5, C/EBP plus HNF4, C/EBP) could not only provide a strong liver-directed gene expression, but also exhibit a high ratio of liver cell to non-livercelloriginreportergeneexpression.Furthermore,mixed ApoB/OTC cassette constructs did not exhibit any significant orientation effect related to the order of ApoB/OTC subcassettes (Figure 3; EGFP expression levels obtained with p1 × ApoB/OTC-GreeN vs p1 × OTC/ApoB-GreeN and p2 × ApoB/OTC-GreeN vs p2 × OTC/ApoB-GreeN).
In sharp contrast to all constructs made up of “natural” composite enhancer elements of the ApoB or the OTC gene, constructs encoding single TFBS of liver-specific transcription factors (p1 × HNF1-GreeN, p3 × HNF1-GreeN, p1 × HNF3-GreeN,p1 × HNF4-GreeN,p1 × HNF5-GreeN, p1 × C/EBP-GreeN) did not achieve EGFP expression levels much higher than background levels (data not shown). Therefore, neither liver-directed nor strong reporter gene expression could be obtained with such single TFBS encoding constructs, demonstrating the superiority of “natural” composite enhancer element constructs.
Since transcriptional control unit constructs encoding ApoB cassettes could exhibit high relative EGFP levels (in our first round of testing by FACS analysis: 4 × ApoB construct reaching nearly up to 70% of expression obtained with the strong hCMV IE driven control plasmid pEGFP-C1; FACS analysis data is depicted in upper panel of Figure 3), we additionally performed a more precise second round testing and also analyzed synthetic ApoB transcriptional control unit transcriptional activities by semi-quantitative RT-PCR and Western blotting (Figures 4 and 5).
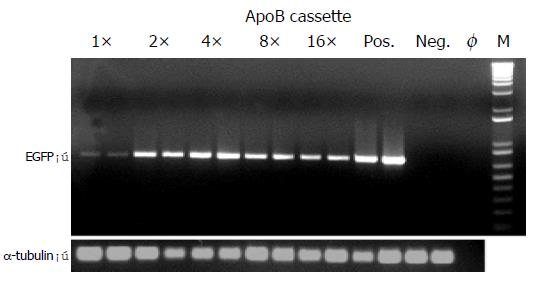
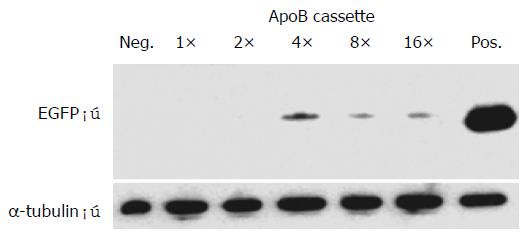
For this purpose, Hep3B cells were transiently transfected with enhancer module plasmids exhibiting single or multimeric ApoB cassettes. Following total RNA extraction and reverse transcription, EGFP expression was analyzed by semi-quantitative PCR analysis which showed a specific band of 717 bp (Figure 4; EGFP). As an internal standard, the α-tubulin house-keeping gene was amplified in parallel giving rise to a specific band of 208 bp (Figure 4; α-tubulin). As a result, a maximum EGFP signal was obtained with the p4 × ApoB-GreeN construct (Figure 4; lanes 5, 6), which corresponded with the FACS analysis data and also exhibited the strongest EGFP expression level for Hep3B cells transfected with the p4 × ApoB-GreeN construct (Figure 3; upper panel, 4 × ApoB, orange bar).
Furthermore, EGFP protein contents obtained in cell lysates from Hep3B cells transiently transfected with enhancer module plasmids exhibiting single or multimeric ApoB cassettes were analyzed by Western blotting (Figure 5), using α-tubulin protein expression as an internal standard. As a result, a maximum EGFP signal was again obtained with the p4 × ApoB-GreeN construct (Figure 5; lane 4), which again corresponded with the FACS analysis data and also exhibited the strongest EGFP expression level for Hep3B cells transfected with the p4 × ApoB -GreeN construct (Figure 3; upper panel, 4 × ApoB, orange bar). Similar results were obtained under neomycin(G418)-selection of Hep3B cells transfected with the p4 × ApoB-GreeN construct, again demonstrating potent expression of the neor/neomycin aminoglycoside phosphotransferase coding sequence driven by the 4 × ApoB-TK-37 promoter cassette (data not shown).
Taken together, semi-quantitative RT-PCR, Western blotting analysis, and G418-selection of Hep3B cells demonstrated maximum transcriptional activities for the multimeric 4 × ApoB cassette construct (p4 × ApoB-GreeN), which fully complied with the data obtained by simple FACS analysis. Importantly, this finding could underline the possibility to first perform simple and quick FACS screening analysis based on our new EGFP/neor fusion reporter gene before doing any detailed analysis on a direct reporter mRNA or protein level.
Proper transcriptional regulation of therapeutic transgene expression constitutes a major requirement in liver-directed gene therapy. Gene regulatory elements used in liver gene therapy should meet several requirements. First, they should drive expression of the therapeutic gene at a sufficient high level. Second, they should restrict expression to the liver. Third, they should be well characterized and have suitable size to allow a most versatile usage in the context of a variety of different gene vector systems. Fourth, the well-known phenomenon of interaction of viral vector sequences with internal liver-specific promoters[24] should be kept at a minimum to avoid any interrelation with the expression of therapeutic transgenes.
Promoters of cellular genes expressed preferentially in the liver have been studied extensively and are used for hepatocyte/hepatoma-specific gene delivery[25-29]. However, the major shortcomings considering their widespread usage in liver gene therapy are the big size (contradictory to the restricted cloning space offered by many vector systems), the low transcriptional activity compared to hCMV or long terminal repeat (LTR) promoter sequences widely used in current gene therapy protocols[30-32], and/or interaction with viral vector sequences influencing expression of therapeutic transgenes[33].
Synthetic promoter/enhancer elements based on minimal promoters offer a promising alternative approach[34]. Combining DNA fragments encoding liver gene transcrip-tion factor binding sites potentially enables both a high degree of structural compactness and a high degree of liver specificity[12,13,35]. Also control elements derived from genes exhibiting a strict liver-restricted regulation like albumin, apolipoproteins, and alpha-1-antitrypsin can be used for this purpose[36,37]. In addition, this synthetic approach helps to exclude any repressor coding sequences preventing unwanted interactions of endogenous repressor factors with TFBS encoded in naturally originating promoter/ enhancer elements.
In an attempt to construct promoters of high liver specificity and activity, we selected hepatospecific transcriptional sequences using the following criteria: short size, wellknown TFBS, expression independent of tight metabolic or hormonal control, and non-species-specificity. We first set out to evaluate both therapeutic efficiency and tissue specificity of this approach in our tissue culture model systems in order to subsequently pre-select liver-specific expression cassettes which could finally be tested within the context of different viral vectors (e.g., adenoviral or lentiviral vectors) both in vitro and in vivo as effective new liver gene transfer systems.
For this purpose, it worked out to be quite helpful to use a newly generated bifunctional EGFP/neor fusion gene cassette, which proved its flexible usage both for transcript quantification by simple FACS analysis (Figure 3) and for selection of stable cell clones (data not shown). Thereby, a proof of principle could be provided demonstrating the possiblity to first perform widespread simple and quick FACS screening analysis based on our new EGFP/neor fusion reporter gene before doing any detailed analysis on a direct reporter mRNA or protein level (Figures 4 and 5). This new approach is of great help in screening any of the numerous modifications of yet available or newly designed promoter cassette constructs.
As a leading result, our comparative analysis of transcriptional activities of synthetically generated promoter cassettes showed that it was not possible to achieve liver-directed gene expression fully approaching that of strong viral promoters such as the hCMV IE promoter. We chose the strong hCMV IE promoter as a reference promoter since it is believed to be similarly active in most tissues at least when used for short term expression. Of the different synthetic promoter cassette configurations tested, the most efficient construction was a multimeric 4 × ApoB cassette construct (p4 × ApoB-GreeN construct) as evidenced by FACS quantification (Figure 3), semi-quantitative RT-PCR (Figure 4), and Western blotting (Figure 5). These results clearly demonstrate that our multimeric 4 × ApoB cassette is a novel regulatory module that allows for an efficient and specific expression of therapeutic transgenes in liver cells. Of particular importance is the small size of this module, which makes it possible to accommodate this transcriptional control unit in any type of viral or non-viral vector, even in conjunction with large effector genes. Therefore, the 4 × ApoB cassette potentially represents a helpful new tool to explore the feasability of liver-directed gene therapy in further detail.
A further advantage of our synthetic minimal promoter based expression cassettes is the possibility to enable finely adjustable levels of therapeutic proteins (low to high) to be delivered/expressed by choosing a 1 × ApoB cassette or a 4 × ApoB cassette. Thereby, increasing or decreasing the number of multimeric monotypic or mixed TFBS-encoding cassettes can adjust the level of therapeutic transgene expression.
In contrast to the good results obtained with “naturally” derived composite enhancer elements from liver-specifically expressed genes, our results with single binding sites of liver-specific transcription factors inserted upstream of the EGFP/neor fusion gene cassette are disappointing (data not shown). This might reflect the strict necessity for optimal spacing of TFBS, a feature that is neglected when single binding sites of liver-specific transcription factors are coupled together in a simple head-to-tail manner.
In comparison to promoters of viral origin, synthetic minimal promoter-based expression cassettes should not undergo in vivo shutdown as a result of DNA methylation processes or because of the presence of transcription factor binding sites at which cognate proteins are not expressed in the quiescent liver[38,39], thereby enabling long-term gene expression. Furthermore, usage of liver-specific promoters in the context of adenoviral vectors has been demonstrated to prevent the generation of antibodies against therapeutic transgenes[40], an eventuality that may occur when a ubiquitous promoter (e.g., hCMV IE) is used. Thus, our synthetic minimal promoter-based expression cassettes might also be helpful in focusing the therapeutic gene expression to the liver concurrently avoiding induction of any significant host immune response to the transgenic protein of choice, thereby prolonging transgene expression in vivo as demonstrated recently for liver-specific gutless adenoviral vectors[41].
In conclusion, we have identified a number of liver-specific promoter sequences that may be valuable tools for targeting heterologous genes to the liver when acceptable levels of therapeutic proteins need to be produced. For reasons of expression efficiency, vector security (especially in the context of suicide gene or other cytotoxic gene expression), and prevention of immune system-induced destruction of transduced cells, our synthetic liver-directed transcriptional control units seem to be a good choice for expression of potential therapeutic genes in the liver. Also clinical usage of non-viral constructs encoding these synthetically generated transcriptional cassettes might be facilitated. This view is based on the recent progress in techniques for delivery of naked DNA to the human liver as well as on their easier high-scale production and better acceptance by regulatory boards than the currently used viral vectors.
The authors thank Drs. M. Spiegel, C.D. Groß, F. Prinz, U. Dau, and S. Lambrecht, former members of the Department of Internal Medicine I, Medical University Clinic Tübingen, for helpful discussions and excellent contributions.
Science Editor Wang XL and Zhu LH, Guo SY Language Editor Elsevier HK
| 1. | Xia D, Zhang MM, Yan LN. Recent advances in liver-directed gene transfer vectors. Hepatobiliary Pancreat Dis Int. 2004;3:332-336. [PubMed] |
| 2. | Prieto J, Herraiz M, Sangro B, Qian C, Mazzolini G, Melero I, Ruiz J. The promise of gene therapy in gastrointestinal and liver diseases. Gut. 2003;52 Suppl 2:ii49-ii54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Shiratori Y, Kanai F, Ohashi M, Omata M. Strategy of liver-directed gene therapy: present status and future prospects. Liver. 1999;19:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Tenenbaum L, Lehtonen E, Monahan PE. Evaluation of risks related to the use of adeno-associated virus-based vectors. Curr Gene Ther. 2003;3:545-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 5. | Schagen FH, Ossevoort M, Toes RE, Hoeben RC. Immune responses against adenoviral vectors and their transgene products: a review of strategies for evasion. Crit Rev Oncol Hematol. 2004;50:51-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Ferry N, Heard JM. Liver-directed gene transfer vectors. Hum Gene Ther. 1998;9:1975-1981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 62] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Ghosh SS, Takahashi M, Thummala NR, Parashar B, Chowdhury NR, Chowdhury JR. Liver-directed gene therapy: promises, problems and prospects at the turn of the century. J Hepatol. 2000;32:238-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Follenzi A, Sabatino G, Lombardo A, Boccaccio C, Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum Gene Ther. 2002;13:243-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 194] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 9. | Kim JW, Wang XW. Gene expression profiling of preneoplastic liver disease and liver cancer: a new era for improved early detection and treatment of these deadly diseases? Carcinogenesis. 2003;24:363-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Suriawinata A, Xu R. An update on the molecular genetics of hepatocellular carcinoma. Semin Liver Dis. 2004;24:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Wu CH, Sapozhnikov E, Wu GY. Evaluation of multicomponent non-viral vectors for liver directed gene delivery. J Drug Target. 2002;10:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 12. | Gehrke S, Jérôme V, Müller R. Chimeric transcriptional control units for improved liver-specific transgene expression. Gene. 2003;322:137-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: designing expression cassettes for gene therapy. Curr Gene Ther. 2004;4:89-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 14. | Paulweber B, Sandhofer F, Levy-Wilson B. The mechanism by which the human apolipoprotein B gene reducer operates involves blocking of transcriptional activation by hepatocyte nuclear factor 3. Mol Cell Biol. 1993;13:1534-1546. [PubMed] |
| 15. | Metzger S, Halaas JL, Breslow JL, Sladek FM. Orphan receptor HNF-4 and bZip protein C/EBP alpha bind to overlapping regions of the apolipoprotein B gene promoter and synergistically activate transcription. J Biol Chem. 1993;268:16831-16838. [PubMed] |
| 16. | Shachter NS, Zhu Y, Walsh A, Breslow JL, Smith JD. Localization of a liver-specific enhancer in the apolipoprotein E/C-I/C-II gene locus. J Lipid Res. 1993;34:1699-1707. [PubMed] |
| 17. | Nishiyori A, Tashiro H, Kimura A, Akagi K, Yamamura K, Mori M, Takiguchi M. Determination of tissue specificity of the enhancer by combinatorial operation of tissue-enriched transcription factors. Both HNF-4 and C/EBP beta are required for liver-specific activity of the ornithine transcarbamylase enhancer. J Biol Chem. 1994;269:1323-1331. [PubMed] |
| 18. | Wu KJ, Wilson DR, Shih C, Darlington GJ. The transcription factor HNF1 acts with C/EBP alpha to synergistically activate the human albumin promoter through a novel domain. J Biol Chem. 1994;269:1177-1182. [PubMed] |
| 19. | Baniahmad A, Steiner C, Köhne AC, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 367] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 20. | Kekulé AS, Lauer U, Weiss L, Luber B, Hofschneider PH. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 277] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 21. | Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2323] [Cited by in RCA: 2329] [Article Influence: 80.3] [Reference Citation Analysis (0)] |
| 22. | Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 391] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 23. | Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858-3863. [PubMed] |
| 24. | Ye X, Liang M, Meng X, Ren X, Chen H, Li ZY, Ni S, Lieber A, Hu F. Insulation from viral transcriptional regulatory elements enables improvement to hepatoma-specific gene expression from adenovirus vectors. Biochem Biophys Res Commun. 2003;307:759-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 25. | Kuriyama S, Yoshikawa M, Ishizaka S, Tsujii T, Ikenaka K, Kagawa T, Morita N, Mikoshiba K. A potential approach for gene therapy targeting hepatoma using a liver-specific promoter on a retroviral vector. Cell Struct Funct. 1991;16:503-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933-10938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 567] [Cited by in RCA: 592] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 27. | Tomizawa M, Yu L, Wada A, Tamaoki T, Kadomatsu K, Muramatsu T, Matsubara S, Watanabe K, Ebara M, Saisho H. A promoter region of the midkine gene that is frequently expressed in human hepatocellular carcinoma can activate a suicide gene as effectively as the alpha-fetoprotein promoter. Br J Cancer. 2003;89:1086-1090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Lu SY, Sui YF, Li ZS, Pan CE, Ye J, Wang WY. Construction of a regulable gene therapy vector targeting for hepatocellular carcinoma. World J Gastroenterol. 2003;9:688-691. [PubMed] |
| 29. | Shi YJ, Gong JP, Liu CA, Li XH, Mei Y, Mi C, Huo YY. Construction of a targeting adenoviral vector carrying AFP promoter for expressing EGFP gene in AFP-producing hepatocarcinoma cell. World J Gastroenterol. 2004;10:186-189. [PubMed] |
| 30. | Ponder KP, Dunbar RP, Wilson DR, Darlington GJ, Woo SL. Evaluation of relative promoter strength in primary hepatocytes using optimized lipofection. Hum Gene Ther. 1991;2:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 31. | Hafenrichter DG, Wu X, Rettinger SD, Kennedy SC, Flye MW, Ponder KP. Quantitative evaluation of liver-specific promoters from retroviral vectors after in vivo transduction of hepatocytes. Blood. 1994;84:3394-3404. [PubMed] |
| 32. | Hafenrichter DG, Ponder KP, Rettinger SD, Kennedy SC, Wu X, Saylors RS, Flye MW. Liver-directed gene therapy: evaluation of liver specific promoter elements. J Surg Res. 1994;56:510-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 33. | Wu X, Holschen J, Kennedy SC, Ponder KP. Retroviral vector sequences may interact with some internal promoters and influence expression. Hum Gene Ther. 1996;7:159-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Xu ZL, Mizuguchi H, Ishii-Watabe A, Uchida E, Mayumi T, Hayakawa T. Optimization of transcriptional regulatory elements for constructing plasmid vectors. Gene. 2001;272:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 130] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 35. | Kramer MG, Barajas M, Razquin N, Berraondo P, Rodrigo M, Wu C, Qian C, Fortes P, Prieto J. In vitro and in vivo comparative study of chimeric liver-specific promoters. Mol Ther. 2003;7:375-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 36. | De Simone V, Cortese R. Transcriptional regulation of liver-specific gene expression. Curr Opin Cell Biol. 1991;3:960-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Noda C, Ichihara A. Regulation of liver-specific gene expression. Cell Struct Funct. 1993;18:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Kay MA, Baley P, Rothenberg S, Leland F, Fleming L, Ponder KP, Liu T, Finegold M, Darlington G, Pokorny W. Expression of human alpha 1-antitrypsin in dogs after autologous transplantation of retroviral transduced hepatocytes. Proc Natl Acad Sci USA. 1992;89:89-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 177] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 39. | Löser P, Jennings GS, Strauss M, Sandig V. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J Virol. 1998;72:180-190. [PubMed] |
| 40. | Pastore L, Morral N, Zhou H, Garcia R, Parks RJ, Kochanek S, Graham FL, Lee B, Beaudet AL. Use of a liver-specific promoter reduces immune response to the transgene in adenoviral vectors. Hum Gene Ther. 1999;10:1773-1781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 131] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 41. | Wang L, Hernández-Alcoceba R, Shankar V, Zabala M, Kochanek S, Sangro B, Kramer MG, Prieto J, Qian C. Prolonged and inducible transgene expression in the liver using gutless adenovirus: a potential therapy for liver cancer. Gastroenterology. 2004;126:278-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |