Published online Sep 7, 2005. doi: 10.3748/wjg.v11.i33.5151
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: September 7, 2005
AIM: A few studies have applied genomic-wide gene expression analysis in inflamed colon tissue sample in ulcerative colitis (UC). We reported the first study of non-inflamed mucosal gene expression in UC and explored its clinical implication.
METHODS: Non-inflamed mucosa was obtained from 6 UC patients who received total colectomy. The gene expression of UC in noninflamed mucosa was monitored with a microarray. For a selected gene, RT-PCR was performed to verify array results and to further examine expression pattern in inflamed mucosa of UC, colorectal cancer tissue and normal mucosa using additional matched pairs.
RESULTS: Two genes showing 2.5-fold decreased expression with significance set at in UC samples were homeo box a4 (HOXa4) and mads box transcription enhancer factor 2, polypeptide B (MEF2b). RT-PCR showed that MEF2b expression in non-inflamed mucosa was significantly downregulated, whereas the expression of MEF2b increased in accordance with the severity of mucosal inflammation. HOXa4 expression both in inflamed and non-inflamed mucosa in UC was consistently downregulated, and the downregulation of HOXa4 was also found in colorectal carcinoma.
CONCLUSION: It is suggested that the MEF2b expression in UC which increase in accordance with inflammation may be induced by the inflammatory mediator. In contrast the downregulation of HOXa4 may be partly involved in the pathogenesis of disease including UC and UC-related carcinogenesis.
- Citation: Toiyama Y, Mizoguchi A, Kimura K, Araki T, Yoshiyama S, Sakaguchi K, Miki C, Kusunoki M. Persistence of gene expression changes in noninflamed and inflamed colonic mucosa in ulcerative colitis and their presence in colonic carcinoma. World J Gastroenterol 2005; 11(33): 5151-5155
- URL: https://www.wjgnet.com/1007-9327/full/v11/i33/5151.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i33.5151
Ulcerative colitis (UC) and Crohn’s disease (CD), two common inflammatory bowel diseases (IBDs) with shared clinical and demographic characteristics, harbor key differences in tissue damage and prognosis that suggest distinctive etiopathogeneic processes. In UC, inflammation with crypt abscess formation is limited to mucosa, while in CD, transmural granulomatous inflammation leads to fibrostenotic lesions and fistula formation[1]. Furthermore, these diseases have complicated clinical manifestations in which genetic, environmental, and microbial factors interact to determine the susceptibility response of immune and nonimmune cellular systems mediating inflammation[2].
In many murine models of colitis recently generated by gene targeting, bacterial flora in the gut appears to be a prerequisite for the development of acute and chronic inflammation[3]. Of great interest, although normal intestinal epithelial cell are hypo-responsive to normal flora in vivo, pathogenic bacteria and bacterial components can initiate an inflammatory response in colitis[4]. These reports suggest that UC mucosa is already a different gene expression compared to normal mucosa without inflammatory change.
Genome-wide gene expression analysis of tissue samples from affected and normal individuals can illuminate important events involved in disease pathogenesis. Recently, several research articles on IBD using cDNA or oligonucleotide array-based technology have been published[5-8]. But previous studies of genomic alteration of IBD-related genes using microarray were directed to inflamed and non-inflamed UC mucosa.
In this study, we first applied this approach of comparing gene expression in non-inflamed UC mucosa to normal human colonic mucosa and searched for these specific changes in gene expression pattern of inflamed UC mucosa and colorectal carcinoma.
This study included 6 UC patients who received total proctocolectomy with ileal J pouch anal anastomosis because of severe distal colitis. The ages of the patients ranged from 20 to 58 years (average 36.1 years). Classic clinical, radiologic, endoscopic, and histopathological criteria were used to confirm all diagnoses[9]. For UC sample, specimens of macroscopically noninflamed proximal colonic mucosa such as cecum or ascending colon was obtained.
Histologically normal samples (six colonic mucosa) were obtained from 6 patients ranging from 40 to 68 years of age (average 53 years) undergoing bowel resection for adenocarcinoma (n = 4), diverticulitis (n = 1), mucosal prolapse syndrome (n = 1). Tissues used for control RNA extraction were obtained from at least 10 cm of the area of pathology and all were histologically normal. Use of patient material was in accordance with Institutional Review Board guidelines and protocol.
All mucosa were dissected from underlying tissue and homogenized in Physcotron (NITI-ON, Japan) for the isolation of total RNA. Total RNA was extracted with Sepasol-RNA (Nacalai Tesque, Tokyo, Japan) and RNA quality was confirmed by agarose gel electrophoresis.
cDNA targets for hybridizations were synthesized by RT from 20 μg/slide of UC- and normal colonic mucosa - total RNA samples with Superscript II RT (Invitrogen) according to the manufacturer’s instructions. AA-dUTP labeled cDNA purified using QIAquick PCR purification Kit (28014:Qiagen). Each AA-dUTP labeled cDNA was coupled with either monoreactiveCy3 (PA23001 Amersham Bioscience) or Cy5 (PA25001 Amersham Bioscience) and incubated for 1 h at 40 °C. Free dye was removed using P30 column (732-6223 BioRad). The separately synthesized Cy3- and Cy5-labeled targets was mixed with 15 μL of Human Cot-1 DNA (15279: Invitrogen), 1 μL of poly A (POLYA GF Invitrogen) and 0.5 μL of t RNA, Yeast (15401-011: Invitrogen) and the mixture was applied to Microcon-YM30 (43409 Millipore) and concentrated to 5-10 μL.
Diluted dye-labeled, 12.5 μL of 20×SSC, 2.5 μL of 10% SDS, 4 μL of 50× Denhardt’s solution, 10 μL of hybridization solution (supplied with AceGene®: Hitachi Software Engineering Co., Ltd) and adequate sterile distilled water were added and concentrated to 44.5 μL. This solution was incubated for 2 min at 95 °C, and cooled on ice, and 0.5 μL of salmon sperm DNA and 5 μL of formamide was added to the solution, and incubated for 5 min at 42 °C. The target cDNA solution was applied directly to the AceGene® Human Oligo Chip (Hitachi Software Engineering Co., Ltd), and coverglass (24 mm×60 mm) was set. Hybridization was performed for 16 h at 42 °C. After hybridization, the slides were washed and rinsed. Finally they were dried with air-spray.
Fluorescent signals were detected on a confocal laser scanner (HB GeneArray Scanner, Affymetrix) and analyzed with the DenasisArray (Hitachi Software Engineering) and Excel (Microsoft Redmond, CA). To eliminate data with low reliability, genes with each hybridization intensity = 0 and each average intensity {(Cy3-Cy3 background)+(Cy5-Cy5 background)<2000} were excluded from all data sets. In addition, genes with average intensity ratio that showed a >2- or 0.5-fold change were selected. In the remaining genes, 2-fold average intensity ratio genes with each intensity ratio<1 on array tests and 0.5-fold average intensity ratio genes with each intensity ratio >1 were excluded. Finally, selected genes were further analyzed for statistical comparison. Mann-Whitney U-test was used, with significance set at P<0.05.
Total RNA was extracted from an additional 6 UC patients’ paired non-inflamed mucosa and inflamed mucosa as well as 9 patients’ paired colon cancer and histological normal colonic mucosa with Sepasol-RNAI. For cDNA synthesis, 2 μg of total RNA was reverse transcribed with Superscript II. RT-PCR was done using specific primers for the array-selected gene. Primers are described in Table 1. Optimum cycling parameters, in the linear range of amplification, consisted of 30 s of denaturation at 94 °C, 30 s of annealing at 58 °C, and 2 min of elongation at 72 °C, and 28 cycles were performed for the selected gene. A control PCR was also done for G3PDH which served as a standard for sample normalization for 25 cycles. Amplified products were separated electrophoretically, visualized, and photographed under UV light after ethidium bromide staining. Gel density was calculated with Sicon image (downloaded sicon’s web site: http://www.siconcorp.com) and gene expressions were quantified. For statistical comparison Mann-Whitney U-test was used, with significance set at P<0.05 to compare differences in the level of gene expression.
| Gene | 5’-Forward | 3’-Reverse |
| Homeo box a4 | ATGACCATGAGCTCGTTTTTGAT | TATGGAGGAGGGAACGGGTG |
| Mads box transcription factor 2b | ATGGGGAGGAAAAAAATCCAGA | CTGTTGGGTCTTCTCTGAAGA |
| G3PDH | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA |
To investigate gene expression profiles of the colon in UC patients, we used AceGene® Human Oligo Chip arrays containing probes for approximately 20 000 human genes. Low reliability data was eliminated by performing data trimming as described in Materials and methods section. After data trimming, the remaining genes were analyzed for statistical comparison between the average intensity of non-inflamed UC group and normal control.
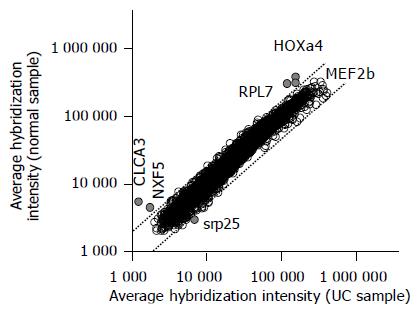
Table 2 lists the gene transcripts displaying a two fold or higher increase or decrease in expression level that was significant at the P<0.05 level. Five gene transcripts displayed a 2.1-4.4-fold lower expression in the non-inflamed UC samples compared with the normal colon samples, whereas only one transcript displayed a 2.3-fold higher expression in the non-inflamed UC samples (Figure 1). Among the six genes, the difference in the expression of the two genes homeo box a4 (NM_002141) and mads box transcription enhancer factor 2 (NM_005919) were statistically significant (P<0.01).
| Accession number | Gene name | AI in controls | AI in UC | Control/UC | P |
| NM_002141 | Homeo box a4 (Hoxa4) | 356 193.4 | 144 705.0 | 2.46 | 0.005 |
| NM_005919 | Mads box transcription enhancer factor 2, polypeptide b (MEF2b) | 283 291.9 | 113 174.8 | 2.5 | 0.009 |
| XM_001566 | Calcium activated chloride channel 3 precursor (CLCA3) | 5 308.8 | 1 188.9 | 4.46 | 0.01 |
| NM_032946 | Nuclear rna export factor 5 (NXF5) | 4 416.7 | 1 681.3 | 2.63 | 0.014 |
| NM_022061 | Ribosomal protein l17 isolog (RPL7) | 299 317.9 | 143 755.9 | 2.08 | 0.027 |
| NM_016638 | Srp25 nuclear protein (SRP25) | 2 872.1 | 6 576.4 | 0.43 | 0.043 |
To verify array results, expression of two genes at the P<0.01 as differentially expressed by oligonucleotide array analysis was examined by RT-PCR using six additional pairs (noninflamed UC mucosa and normal control).
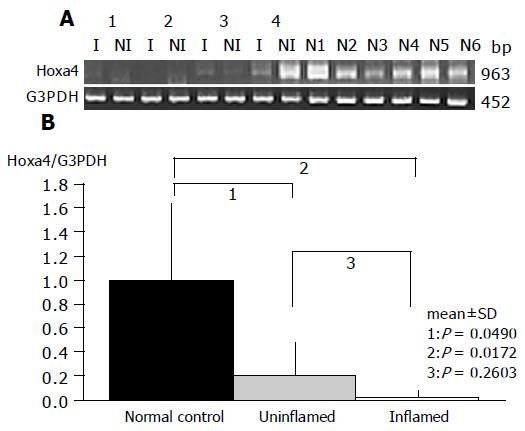
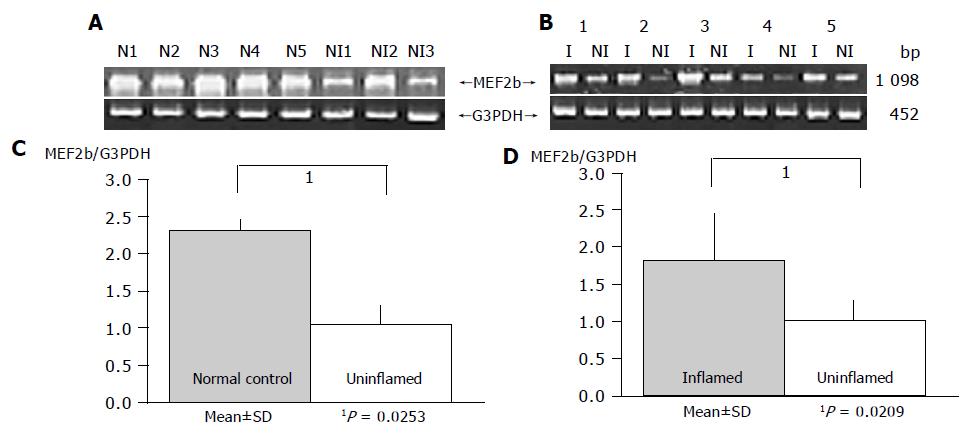
As a result, the expression of HOXa4 and MEF2b in non-inflamed UC mucosa was significantly reduced compared to that in the normal mucosa (P = 0.0490 and P = 0.0253, respectively), which gave results consistent with those by oligonucleotide array analysis (Figures 2 and 3).
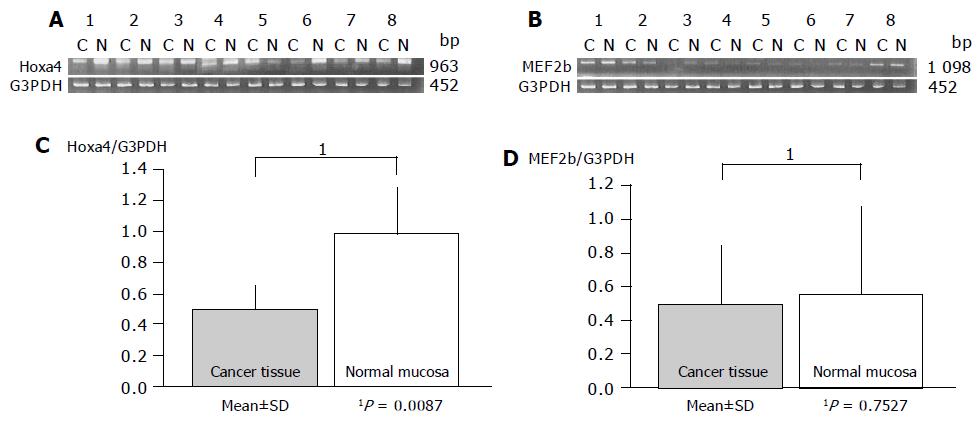
Gene expression patterns of both these genes obtained from inflamed UC mucosa were examined comparing them with the non-inflamed UC mucosa and normal mucosa using RT-PCR. MEF2b expression in inflamed UC mucosa was significantly upregulated compared to that in non-inflamed UC mucosa (P = 0.0209; Figure 3). But there is no difference in MEF2b expression between normal control and inflamed UC mucosa (Figure 3). In contrast, no difference in the expression pattern of HOXa4 was found between those with and without colonic inflammation (P = 0.2603; Figure 2). Furthermore, the expression of HOXa4 in inflamed mucosa was significantly downregulated compared to that in normal mucosa (P = 0.0172; Figure 2). RT-PCR on eight samples of matched colon cancer and normal mucosa showed that the expression of HOXa4 in colon cancer tissue was significantly downregulated in 8 of 8 (100%). But the expression of MEF2b had no difference between colorectal carcinoma and normal mucosa (Figure 4).
Our present study is the first trial which draws a comparison between genomic alternation in non-inflamed UC mucosa and normal human colonic mucosa. This approach has allowed us to examine whether non-inflamed UC mucosa shows a different gene expression compared to normal control mucosa. We showed the intensities of the two genes HOXa4 and MEF2b to be reduced in the non-inflamed mucosa in patients with UC using the cDNA microarray. In addition, we compared the levels of the expression of mRNA of these two genes among control, non-inflamed and inflamed mucosa in UC, and colonic cancer tissues by RT-PCR.
HOXa4 includes the homeo box (HOX) family genes that are functionally homologous to the drosophila HOM-C gene. There are 39 HOX genes in the human genome that are arranged in four clusters on four different chromosomes. They are expressed at the beginning of gastrulation and define the body’s axis formation. In adults, HOX B8 is involved in leukomorphogenesis[10]. Other HOX genes play a role in malignant transformation, through the regulation of cell-cycle checkpoints (e.g. P53) or protooncogenes (e.g. ras[11]). In cancer cells, HOX is either over-expressed (e.g. HOX C4 and HOX C6, in non-Hodgkin’s lymphoma[10]) or downregulated (e.g. HOX B6, C8, C9 in adenocarcinoma and colon cells[12]). In our experiments, HOXa4 gene expression was downregulated in UC mucosa compared to normal mucosa whether colonic inflammation exists or not and this gene expression in colon cancer tissue was also significantly lower than normal mucosa with additional RT-PCR.
Earlier studies showed that HOX A4 is regulated both by retinoic acid and the ras oncogene[13]. Retinoic acid increased HOX A4 expression and guided the cells toward differentiation, while the ras oncogene decreased HOX A4 expression and guided the cells toward proliferation. Burmer previously reported that UC-associated neoplasia has a low incidence of K-ras mutations[14], but Ierardi et al, showed that the persistently increased epithelial proliferation associated with ras p21 expression in UC may be due to the action of an abnormal, mutated ras gene that could play a role in UC-related tumorigenesis[15]. Steingart et al[16], previously reported that SNH (a lipophilic analog of vasoactive intestinal peptide (VIP)) increased the expression of HOX A4 and reduced Oct-3 mRNA level and demonstrated inhibition of HT 29 colon cancer cell line proliferation by SNH.
This may suggest that decreased HOXa4 gene expression partly explains the pathogenesis of UC and UC-related carcinogenesis without ras protein participation and SNH might serve as a lead compound to prevent colitic cancer for UC patients who have received long-term medical therapy.
MEF2 (myocyte enhancer factor-2) was first described as a muscle-enriched transcription factor that bound to an A/T-rich DNA sequence in the control regions of numerous muscle-specific genes[17]. There are four human MEF2 genes, MEF2A, MEF2B, MEF2C and MEF2D, which are expressed in distinct, but overlapping patterns during embryogenesis and in adult tissues[18]. Recent studies have revealed a central role for MEF2 family of transcription factors in linking calcium-dependent signaling pathways to the genes responsible for cell division, differentiation and apoptosis[19]. In regulation of cell proliferation, it was made with the discovery that MEF2 regulates serum-inducible expression of c-jun, which positively regulates cell-cycle progression[20]. MEF2 has since been shown to play a role in the induction of the c-jun promoter in response to signals emanating from multiple cell surface receptors, including G-protein-coupled receptors[21], the epidermal growth factor receptor[22], the lipopolysaccharide receptor[23], and the CD28 co-stimulatory receptor in T lymphocytes[24]. It is therefore, conceivable that constant decreased expression of MEF2 might regulate epithelial cell proliferation and cell-cycle progression and result in difficulty of epithelial cell regeneration in noninflamed UC mucosa.
Interestingly, the present study demonstrated that MEF2 was overexpressed in inflamed UC mucosa compared to non-inflamed UC mucosa using RT-PCR. MEF2 stimulates the expression of the gene encoding the orphan nuclear hormone receptor Nur77[25], which is a potent activator of cytochrome c-mediated apoptosis. The present results suggest that ulceration of mucosa might be related to epithelial cell apoptosis induced by MEF2 overexpression in inflamed UC mucosa.
In conclusion, the present study indicates that UC mucosa already has a different gene expression compared to normal mucosa without inflammatory change. Downregulated HOXa4 expression pattern in UC mucosa consistently as well as in colorectal carcinoma may play an important role for carcinogenesis in UC mucosa. Since MEF2 expression pattern correlated to inflammation, it is a key molecule for colonic epithelial cell apoptosis in inflamed mucosa and cell remodeling difficulty in non-inflamed UC mucosa. In a subsequent research, we hope to study further to identify the function of selected UC-related gene.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Kinsner J, Shorter R. Infalmmatory Bowel disease, 5th edn.Lea&Febiger, Philadelphia, PA. 1999;. |
| 2. | Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1493] [Cited by in RCA: 1485] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 3. | Hibi T, Ogata H, Sakuraba A. Animal models of inflammatory bowel disease. J Gastroenterol. 2002;37:409-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Savkovic SD, Koutsouris A, Hecht G. Activation of NF-kappaB in intestinal epithelial cells by enteropathogenic Escherichia coli. Am J Physiol. 1997;273:C1160-C1167. [PubMed] |
| 5. | Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1-11. [PubMed] |
| 6. | Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn's disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 292] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 7. | Uthoff SM, Eichenberger MR, Lewis RK, Fox MP, Hamilton CJ, McAuliffe TL, Grimes HL, Galandiuk S. Identification of candidate genes in ulcerative colitis and Crohn's disease using cDNA array technology. Int J Oncol. 2001;19:803-810. [PubMed] |
| 8. | Masuda H, Takahashi Y, Asai S, Takayama T. Distinct gene expression of osteopontin in patients with ulcerative colitis. J Surg Res. 2003;111:85-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Truelove SC, Witts LJ. Cortisone in ulcerative colitis; final report on a therapeutic trial. Br Med J. 1955;2:1041-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1832] [Cited by in RCA: 1867] [Article Influence: 26.7] [Reference Citation Analysis (1)] |
| 10. | Cillo C, Faiella A, Cantile M, Boncinelli E. Homeobox genes and cancer. Exp Cell Res. 1999;248:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 122] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 174] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Vider BZ, Zimber A, Chastre E, Gespach C, Halperin M, Mashiah P, Yaniv A, Gazit A. Deregulated expression of homeobox-containing genes, HOXB6, B8, C8, C9, and Cdx-1, in human colon cancer cell lines. Biochem Biophys Res Commun. 2000;272:513-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Doerksen LF, Bhattacharya A, Kannan P, Pratt D, Tainsky MA. Functional interaction between a RARE and an AP-2 binding site in the regulation of the human HOX A4 gene promoter. Nucleic Acids Res. 1996;24:2849-2856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Burmer GC, Levine DS, Kulander BG, Haggitt RC, Rubin CE, Rabinovitch PS. c-Ki-ras mutations in chronic ulcerative colitis and sporadic colon carcinoma. Gastroenterology. 1990;99:416-420. [PubMed] |
| 15. | Ierardi E, Principi M, Francavilla R, Passaro S, Noviello F, Burattini O, Francavilla A. Epithelial proliferation and ras p21 oncoprotein expression in rectal mucosa of patients with ulcerative colitis. Dig Dis Sci. 2001;46:1083-1087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Steingart RA, Heldenberg E, Pinhasov A, Brenneman DE, Fridkin M, Gozes I. A vasoactive intestinal peptide receptor analog alters the expression of homeobox genes. Life Sci. 2002;71:2543-2552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 17. | Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 808] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 18. | Breitbart RE, Liang CS, Smoot LB, Laheru DA, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095-1106. [PubMed] |
| 19. | Blaeser F, Ho N, Prywes R, Chatila TA. Ca(2+)-dependent gene expression mediated by MEF2 transcription factors. J Biol Chem. 2000;275:197-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 164] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 20. | Han TH, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907-2915. [PubMed] |
| 21. | Coso OA, Montaner S, Fromm C, Lacal JC, Prywes R, Teramoto H, Gutkind JS. Signaling from G protein-coupled receptors to the c-jun promoter involves the MEF2 transcription factor. Evidence for a novel c-jun amino-terminal kinase-independent pathway. J Biol Chem. 1997;272:20691-20697. [RCA] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065-1073. [PubMed] |
| 23. | Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 631] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 24. | Shin HM, Han TH. CD28-mediated regulation of the c-jun promoter involves the MEF2 transcription factor in Jurkat T cells. Mol Immunol. 1999;36:197-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Woronicz JD, Lina A, Calnan BJ, Szychowski S, Cheng L, Winoto A. Regulation of the Nur77 orphan steroid receptor in activation-induced apoptosis. Mol Cell Biol. 1995;15:6364-6376. [PubMed] |