Published online Sep 7, 2005. doi: 10.3748/wjg.v11.i33.5103
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: September 7, 2005
AIM: To investigate the association between the genetic polymorphisms of ADH2 and ALDH2, lifetime alcohol consumption and esophageal cancer risk in the Taiwanese men.
METHODS: Between August 2000 and June 2003, 134 pathologically-proven esophageal squamous cell carcinoma male patients and 237 male controls were recruited from Kaohsiung Medical University Hospital and Kaohsiung Veterans General Hospital in southern Taiwan. ADH2 and ALDH2 polymorphisms were genotyped using PCR-RFLP.
RESULTS: Compared to those with ADH2*2/*2, individuals with ADH2*1/*2 and ADH2*1/*1 had 2.28- and 7.14-fold, respectively, increased risk of developing esophageal cancer (95%CI = 1.11-4.68 and 2.76-18.46) after adjusting for alcohol consumption and other covariates. The significant increased risk was also noted among subjects with ALDH2*1/*2 (adjusted OR (AOR) = 5.25, 95%CI = 2.47-11.19), when compared to those with ALDH2*1/*1. The increased risk of esophageal cancer was made greater, when subjects carried both ADH2*1/*1 and ALDH2*1/*2, compared to those with ADH2*1/*2 or ADH2*2/*2 and ALDH2*1/*1 (AOR = 36.79, 95%CI = 9.36-144.65). Furthermore, we found a multiplicative effect of lifetime alcoholic consumption and genotypes (ADH2 and ALDH2) on esophageal cancer risk.
CONCLUSION: Our findings suggest that polymorphisms of ADH2 and ALDH2 can modify the influence of alcoholic consumption on esophageal cancer risk.
- Citation: Wu CF, Wu DC, Hsu HK, Kao EL, Lee JM, Lin CC, Wu MT. Relationship between genetic polymorphisms of alcohol and aldehyde dehydrogenases and esophageal squamous cell carcinoma risk in males. World J Gastroenterol 2005; 11(33): 5103-5108
- URL: https://www.wjgnet.com/1007-9327/full/v11/i33/5103.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i33.5103
Epidemiologic studies have demonstrated that drinking alcoholic beverages is associated with the development of cancers in the oral cavity, pharynx, larynx, and esophagus[1]. Our previous study also reported that, in addition to cigarette smoking and areca chewing, alcohol consumption is one of the leading risk factors for esophageal cancer in Taiwan[2]. In that study, we found that subjects who consumed more than 1 220 g/year of alcohol (about 4 cans of beer per day for 20 years) in a lifetime were found to have a 9.7-fold higher risk for esophageal cancer than those who did not drink (95%CI = 4.3-22.0).
When ethanol is consumed through drinking, it is metabolized primarily by class I alcohol dehydrogenase (ADH2) into acetaldehyde, an intermediate metabolite, and then it is metabolized by aldehyde dehydrogenase (ALDH2) into acetic acid in humans[3]. Acetaldehyde, a well-known carcinogen in animals, plays an important role in alcohol toxicity in humans[4].
Genetic variation in the ability to metabolize alcohol might be associated with esophageal cancer risk. ADH2 and ALDH2 genes are located on chromosomes 4q22 and 12q24, respectively. One amino acid has been found to change from arginine (CGC) (ADH2*1) to histidine (CAC) (ADH2*2) in ADH2 gene at codon 47 of exon 3[5]. Bosron and Li[3] has found ADH2*2/*2 to have about 40 times higher Vmax than ADH2*1/*1, because ADH2*2/*2 encodes a superactive subunit of ADH2 in the oxidation of ethanol, causing acetaldehyde to accumulate in the circulation. For ALDH2, a single nucleotide transition from glutamic acid to lysine (G→A) at codon 487 of exon 12 has also been reported[6]. The mutant allele of ALDH2*2 (A/A) has relatively lower ALDH2 activity than ALDH2*1 allele (G/G)[7] in the metabolization of acetaldehyde into acetic acid, leading to the accumulation of circulatory acetaldehyde[8].
Many studies from Japan have reported that subjects with ADH2*1/*1 or ALDH2*1/*2 genotypes were much more susceptible to esophageal cancer than those with the genotypes[9-15]. In Taiwan, however, only one study has reported the relationship between genetic polymorphisms of ADH2 and ALDH2 and esophageal cancer risk[16]. In that study, Chao and his colleagues[16], studying 59 esophageal cancer patients who were also alcoholics and 222 alcoholic controls, found the allele frequencies of ADH2*1 and ALDH2*2 in alcoholic cancer patients to be 51% and 31%, respectively, much higher frequencies than those found in alcoholics without cancer (39%; P<0.025 and 8%; P<0.001). In this study, we further investigated the independent or combined effects of the ADH2 and ALDH2 gene polymorphisms on esophageal cancer risks in the Taiwanese men. We also evaluated interaction of ADH2 and ALDH2 with alcohol consumption and study the interaction’s relationship to esophageal cancer risk.
In this hospital-based case-control study, we recruited male case patients with new histological diagnosis of esophageal squamous cell carcinoma from Kaohsiung Medical University Hospital and Kaohsiung Veterans General Hospital in the southern Taiwan from August 2000 to June 2003. The Department of Preventive Medicine at each hospital chose 1-2 age-matched (±4 years) healthy male controls who had no malignancies to give blood specimens at the time as the cases. In total, 134 cases and 237 controls were interviewed and genotyped from peripheral blood specimens.
Using a standardized questionnaire, trained interviewers collected demographic characteristics and information about cigarette, alcohol and areca consumption from study subjects[2]. Information on habitual substance use included whether the subject had been a habitual areca chewer, cigarette smoker, or an alcoholic in his or her lifetime, what year the subject started and quitted, the duration of consumption and the daily amount consumed, and type of alcoholic beverage consumed. This study was approved by Kaohsiung Medical University Hospital’s IRB. Informed consent was obtained from all subjects.
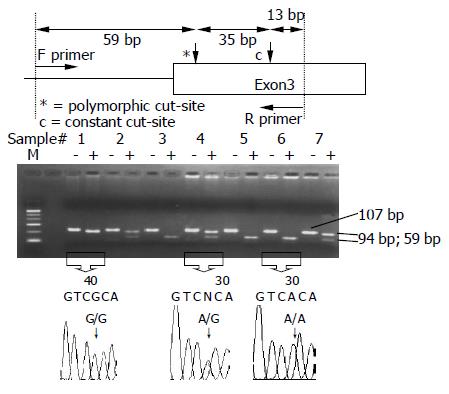
ADH2 polymorphisms at codon 47 were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)[17,18]. The two primers were 5’-AAT CTT TTC TGA ATC TGA ACA G-3’ (upstream) and 5’-GAA GGG GGG TCA CCA GGT TGC-3’ (downstream). The PCR was carried out with 100 ng/μL genomic DNA, 10× buffer, 2.5 mmol/L dNTP, 20 pmol/L of each primer, 50 mmol/L MgCl2 and 5 U Taq polymerase in a 25 μL reaction mixture. The PCR condition was performed by initial denaturation at 95 °C for 5 min, followed by 40 cycles at 95 °C for 1 min, 55 °C for 57.5 s, 72 °C for 45 s and a final extension at 72 °C for 10 min. The final PCR product was digested with MaeIII for 4 h at 55 °C and electrophoresed and viewed on a 3% agarose gel. When a MaeIII restriction site was presented, the 94-bp fragment was digested into two fragments: lengths of 59 and 35 bp. Individuals with ADH2*2/*2 and ADH2*1/*2 had 59- and 35-bp fragments and 94-, 59- and 35-bp fragments, respectively, whereas those with ADH2*1/*1 had only a 94-bp fragment (Figure 1).
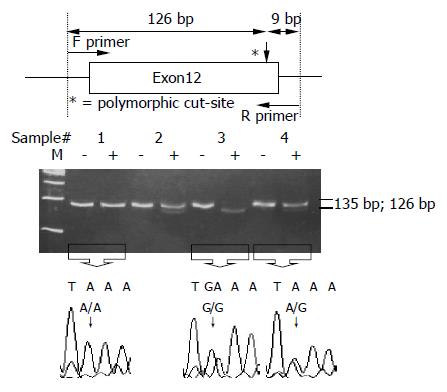
ALDH2 polymorphisms at codon 487 were also determined by PCR-RFLP[11,19]. The DNA sample was amplified with two primers: 5’-CAA ATT ACA GGG TCA ACT GCT-3’ (upstream) and 5’-CCA CAC TCA CAG TTT TCT CTT-3’ (downstream). The PCR was carried out with 100 ng/μL genomic DNA, 10× buffer, 2.5 mmol/L dNTP, 20 pmol/L of each primer, 50 mmol/L MgCl2 and 5 U Taq polymerase in a 25 μL reaction mixture. The PCR condition was performed by initial denaturation at 95 °C for 5 min, followed by 35 cycles at 94 °C for 15 s, 58 °C for 1 min 30 s, 72 °C for 30 s and a final extension at 72 °C for 10 min. The final PCR product was digested with MboII for 16 h at 37 °C and electrophoresed and viewed on a 12% polyacrylamide gel. When a MboII restriction site was presented, the 135-bp fragment was digested into two fragments: lengths of 126 and 9 bp. Individuals with ALDH2*1/*1 and ALDH2*1/*2 had 126- and 9-bp fragments and 135-, 126- and 9-bp fragments, respectively, whereas those with ALDH2*2/*2 had only a 135-bp fragment (Figure 2).
In addition, 20 case patients provided paired samples of tumors and peripheral blood for genotyping. We found that the evidence for both genotypes for ADH2 and ALDH2 were exactly the same in each case patient.
Three different genotypes of each ADH2 and ALDH2 were first sequenced to confirm the findings, then in each genotyping set (~10 samples), we included one positive control and one negative control sample. The positive control sample was included to ensure complete digestion of the PCR product by restriction enzymes. The negative control was placed with the same reagents as those used with actual samples, with the exception of DNA templates. In addition, about 3% of subjects (n = 12/378) were randomly selected for re-genotyping (the results were the same).
Hardy-Weinberg equilibrium statistic was calculated first. Genotype frequencies, demographic characteristics, and substance use (cigarette, alcohol, and areca) in cases and controls were compared by Student’s t, χ2, or Fisher’s exact tests.
For substance use, subjects who reported smoking more than 10 cigarettes/week for at least 6 mo were defined as cigarette smokers, those who reported regularly chewing betel quid for at least 6 mo were defined as betel chewers, and those who reported drinking beer, wine, or distilled spirits more than one time per week for at least 6 mo were defined as alcoholics.
Using multiple logistic regression models, we determined the relationship of ADH2 and ALDH2 polymorphisms with esophageal cancer risk after adjusting for other covariates, including age (continuous variable), educational level (>high school, high school, and <high school), cigarette smoking (yes vs no), alcohol consumption (yes vs no) and areca chewing (yes vs no). Since the interactive effect between lifetime alcohol consumption and ADH2 and ALDH2 polymorphisms on esophageal cancer risk was also examined in this study, lifetime consumption of alcoholic beverage was calculated by multiplying the concentration of alcohol in the consumed beverage by the amount consumed per day by the number of years consumed, resulting in number designated as grams per year. The median cut-off point for lifetime alcohol consumption was 1 500 g/year, which equaled an average of five 300-350 cm3 cans of beer (5% alcohol) per day for 20 consecutive years. Subjects were also classified as non-drinkers if they consumed ≤1 500 g/year, and those who consumed >1 500 g/year were used for the subsequent interaction analysis. The data were analyzed using the SAS 8.1 statistical package; all P-values were two-sided.
We found 231 pathology-proven cases of esophageal squamous cell carcinoma at Kaohsiung Medical University Hospital and Kaohsiung Veterans General Hospital between August 2000 and June 2003. Mean ages (±SD) of the recruited patients (n = 134) and non-recruited patients (n = 97) were 58.9±12.6 and 59.0±13.6 years, respectively, with no significant difference (P = 0.81).
Mean ages (range) of the 134 cases and 237 controls were 58.9 years (34-82 years) and 57.8 years (34-82 years), respectively, with no significant difference (Table 1). Educational level (P<0.0001) and the habitual use of cigarettes (P<0.0001), alcohol (P<0.0001), and areca (P<0.0001) were most significant predictors of esophageal cancer risk. We found cigarette smokers, alcoholics, and areca chewers had a 13.61-, 9.90-, and 13.33-fold higher risk, respectively, for developing esophageal cancer than non-smokers, non-drinkers, and non-chewers (95%CI = 6.81-27.21, 5.89-16.64, and 7.33-24.26, respectively) (data not shown).
| Variables | Cases (%)n = 134 | Controls (%)n = 237 | Crude OR (95%CI) | P |
| Age (yr) | ||||
| Mean±SD | 58.9±12.6 | 57.8±11.8 | – | 0.41 |
| Education (%) | ||||
| <High school | 107 (76.1) | 86 (36.3) | 1 | |
| High school | 23 (17.1) | 94 (39.7) | 0.20 (0.11–0.34) | |
| >High school | 4 (3.0) | 57 (24.1) | 0.06 (0.06–0.16) | <0.0001 |
| Ethnicity (%) | ||||
| Mainlander | 102 (76.1) | 186 (78.5) | 1 | |
| Fukienese | 25 (18.7) | 33 (13.9) | 1.38 (0.78–2.45) | |
| Others | 7 (5.2) | 18 (7.6) | 0.71 (0.29–1.75) | 0.37 |
| Cigarette smoking (%) | ||||
| No | 10 (7.5) | 124 (52.3) | 1 | |
| Yes | 124 (92.5) | 113 (47.7) | 13.61 (6.81–27.21) | <0.0001 |
| Alcohol consumption (%) | ||||
| No | 24 (17.9) | 162 (68.4) | 1 | |
| Yes | 110 (82.1) | 75 (31.6) | 9.9 (5.89–16.64) | <0.0001 |
| Areca chewing (%) | ||||
| No | 66 (49.3) | 220 (92.8) | 1 | |
| Yes | 68 (50.8) | 17 (7.2) | 13.33 (7.33–24.26) | <0.0001 |
The prevalence (number) of ADH2*1/*1, ADH2*1/*2, and ADH2*2/*2 in the controls was 6.8% (16/237), 38.4% (91/237), and 54.9% (130/237), respectively. The gene frequency of the ADH2*1 allele was 25.9%. The distribution of the different genotypes among the 237 controls closely conformed to expected Hardy-Weinberg frequencies (χ2 = 0.00; df = 2; P = 1.00). The prevalence (number) of ALDH2*1/*1, ALDH2*1/*2, and ALDH2*2/*2 in the controls was 50.6% (120/237), 44.3% (105/237), and 5.1% (12/237), respectively. The gene frequency of the ALDH2*2 allele was 27.2%. The distribution of the different genotypes among the 237 controls also closely conformed to expected Hardy-Weinberg frequencies (χ2 = 1.95; df = 2; P = 0.38).
Table 2 shows the association of ADH2 and ALDH2 genotypes with esophageal cancer risk before and after adjusting for other covariates. Subjects with ADH2*1/*2 and ADH2*1/*1 had 2.28 (95%CI = 1.11-4.68)- and 7.14 (95%CI = 2.76-18.46)-fold, respectively, greater risk of developing esophageal cancer than the subjects with ADH2*2/*2, after adjusting for age, educational level, cigarette smoking, alcohol consumption, areca chewing, and ALDH2 polymorphism. In addition, the effect of ADH2*1/*1 was still significantly elevated (adjusted OR (AOR) = 4.80; 95%CI = 2.02-11.44), when compared to the combined ADH2*1/*2 and ADH2*2/*2 (data not shown).
| Genotypes | Cases (%) n = 134 | Controls (%) n = 237 | OR (95%CI) | AOR (95%CI)1 |
| ADH2 | ||||
| *2/*2 | 46 (34.3) | 130 (54.9) | 1 | 1 |
| *1/*2 | 49 (36.6) | 91 (38.4) | 1.52 (0.94–2.47) | 2.28 (1.11–4.68) |
| *1/*1 | 39 (29.1) | 16 (6.8) | 6.89 (3.52–13.49) | 7.14 (2.76–18.46) |
| P for HW2 | 1.00 | |||
| ALDH2 | ||||
| *1/*1 | 32 (23.9) | 120 (50.6) | 1 | 1 |
| *1/*2 | 99 (73.9) | 105 (44.3) | 3.54 (2.19–5.70) | 5.25 (2.47–11.19) |
| *2/*2 | 3 (2.2) | 12 (5.1) | 0.94 (0.25–3.52) | 2.44 (0.44–13.55) |
| P for HW2 | 0.38 |
For ALDH2 genotypes, subjects with ALDH2*1/*2 (AOR = 5.25, 95%CI = 2.47-11.19) were found to have a significantly more increased risk of esophageal cancer compared to those with ALDH2*1/*1 (Table 2). The risk for subjects with ALDH2*2/*2 was slight, but not significantly higher than those with ALDH2*1/*1 (AOR = 2.44; 95%CI = 0.44-13.55). However, compared to ALDH2*1/*1, the combined risk effect of ALDH2*1/*2 and ALDH2*2/*2 was significantly elevated (AOR = 5.20; 95%CI = 2.46-11.00) (data not shown).
Table 3 shows the joint effect of ADH2 and ALDH2 genotypes on esophageal cancer risk. The significantly increased trend for esophageal cancer risk was noted in subjects with ADH2*1/*2 or *2/*2 and ALDH2*1/*1, ADH2*1/*1 and ALDH2*1/*1, ADH2*1/*2 or *2/*2 and ALDH2*1/*2, to ADH2*1/*1 and ALDH2*1/*2. However, the increased risk was not pronounced in a group of ALDH2*2/*2, possibly because of small sample size.
| ADH2 | ALDH2 | Cases (%) n = 134 | Controls (%) n = 237 | OR (95%CI) | AOR (95%CI)1 |
| *1/*2 or *2/*2 | *1/*1 | 23 (17.2) | 112 (47.3) | 1 | 1 |
| *1/*1 | 9 (6.7) | 8 (3.4) | 4.20 (1.49–11.81) | 3.54 (0.93–13.53) | |
| *1/*2 or *2/*2 | *1/*2 | 69 (51.5) | 98 (41.4) | 2.63 (1.58–4.37) | 4.81 (2.17–10.70) |
| *1/*1 | 30 (22.4) | 7 (3.0) | 16.00 (6.40–39.99) | 36.79 (9.36–144.65) | |
| *1/*2 or *2/*2 | *2/*2 | 3 (2.2) | 11 (4.6) | 1.02 (0.27–3.88) | 3.40 (0.63–18.33) |
| *1/*1 | 0 | 1 (0.4) | – | – |
Table 4 shows the effect of a lifetime interaction between alcohol consumption and ADH2 and ALDH2 genotypes on esophageal cancer risk. Using non-drinkers with ADH2*1/*2 or *2/*2 and ALDH2*1/*1 as a baseline, we found that the higher the lifetime consumption of alcohol, the greater the risk for esophageal cancer in each category of ADH2 and ALDH2 genotypes, except for a group of ALDH2*2/*2. When categorizing the subjects into amount of alcohol consumed in a lifetime (non-drinkers, drinkers with ≤1 500 g/year, or drinkers with >1 500 g/year), we found an increased trend for esophageal cancer risk in the more highly susceptible ADH2 and ALDH2 genotypes. Compared to baseline, the risk for developing esophageal cancer increased in subjects carrying ADH2*1/*1 and ALDH2*1/*2 and consuming more than 1 500 g/year of alcohol in a multiplicative fashion (AOR = 139.35, 95%CI = 10.05- , Table 5).
| ADH2 | ALDH2 | Alcohol consumption (g/yr) | Cases (%) n = 134 | Controls (%) n = 237 | OR (95%CI) | AOR (95%CI)1 |
| *1/*2 or *2/*2 | *1/*1 | Non-drinker | 3 (2.2) | 66 (27.9) | 1 | 1 |
| ≤1 500 | 5 (3.7) | 19 (8.0) | 5.79 (1.27-26.46) | 3.76 (0.65-21.69) | ||
| >1 500 | 15 (11.2) | 27 (11.4) | 12.22 (3.27-45.66) | 6.07 (1.46-25.26) | ||
| *1/*1 | *1/*1 | Non-drinker | 0 | 2 (0.8) | – | – |
| ≤1 500 | 4 (3.0) | 4 (1.7) | 22.00 (3.62-133.81) | 14.91 (1.92-115.95) | ||
| >1 500 | 5 (3.7) | 2 (0.8) | 55.00 (7.39-409.22) | 33.54 (3.52-319.89) | ||
| *1/*2 or *2/*2 | *1/*2 | Non-drinker | 13 (9.7) | 76 (32.1) | 3.76 (1.03-13.78) | 2.92 (0.73-11.63) |
| ≤1 500 | 31 (23.1) | 15 (6.3) | 45.47 (12.26-168.68) | 26.67 (6.05-117.57) | ||
| >1 500 | 25 (18.7) | 7 (3.0) | 78.57 (18.83-327.89) | 39.26 (7.05-218.68) | ||
| *1/*1 | *1/*2 | Non-drinker | 5 (3.7) | 6 (2.5) | 18.33 (3.50-96.18) | 18.60 (2.69-128.77) |
| ≤1 500 | 14 (10.5) | 1 (0.4) | 308 (29.80-3 183.05) | 139.35 (10.05-∞) | ||
| >1 500 | 11 (8.2) | 0 | – | – | ||
| *1/*2 or *2/*2 | *2/*2 | Non-drinker | 3 (2.2) | 11 (4.6) | 6 (1.07-33.60) | 2.20 (0.33-14.47) |
| ≤1 500 | 0 | 0 | – | – | ||
| >1 500 | 0 | 0 | – | – | ||
| *1/*1 | *2/*2 | Non-drinker | 0 | 1 (0.4) | – | – |
| ≤1 500 | 0 | 0 | – | – | ||
| >1 500 | 0 | 0 | – | – |
The present study finds alcohol consumption to be the major risk of esophageal cancer in the Taiwanese men. However, ethanol per se is not carcinogenic[1]. Researchers have paid much more attention to the relationship between acetaldehyde, the first metabolite of alcohol, and cancer risk, even though the detailed mechanisms of alcohol-associated cancers, such as esophageal cancer are still puzzling. Ethanol is metabolized mainly by ADH2 to become acetaldehyde and then further metabolized by ALDH2 to form acetic acid. Acetaldehyde is a very active intermediate able to attack DNA or protein to form adducts[20,21]. In addition, Yin and his colleagues[22] have reported the ADH activity in the 15 specimens of esophageal mucosa from the Chinese men to be about four-fold greater than that of gastric mucosa in the same population (n = 7). In contrast, the esophageal ALDH activity is only 20% of that in the stomach, suggesting that the concentration of acetaldehyde in esophagus is possibly much higher than that in the stomach after consuming the beverages.
Since the major metabolic pathway for ethanol is determined by ADH2 and ALDH2, polymorphisms on these two genes may affect the formation of acetaldehyde. For ALDH2 genotypes, Enomoto and his colleagues[23] reported that subjects with ALDH2*1/*2 had an average increase in peak blood acetaldehyde level that was five-fold higher than those with ALDH2*1/*1 after drinking a small amount of ethanol (0.1 g/kg of body weight), though this study did not investigate the effect of ADH2 genotypes on acetaldehyde level. Another study done by Mizoi and his colleagues[24] found that, after consuming beverages, the blood acetaldehyde concentrations in those with ALDH2*2/*2 and ALDH2*1/*2 were 19- and 6-fold higher than in those with ALDH2*1/*1, respectively. The results of these studies suggest that subjects with ALDH2*2/*2 and ALDH2*1/*2 may have a higher risk of developing esophageal cancer than those with ALDH2*1/*1, which is consistent with our findings.
However, our study found no subjects with the highest susceptible genotype of ALDH2, ALDH2*2/*2, to be drinkers. Crabb et al[8], and Thomasson et al[25], found that individuals deficient in ALDH2 activity (ALDH2*2/*2 genotype) can develop intense facial flushing responses, because of high blood acetaldehyde levels, after drinking alcohol. This unpleasant discomfort may prevent people from consuming beverages and may keep them from developing alcoholism, which can explain our findings in this study.
In this study, we found subjects with ADH2*1 allele had a higher risk of developing esophageal cancer than those with ADH2*2 allele. Previous epidemiological studies from Japan also reported a significantly higher risk for esophageal cancer among Japanese drinkers with ADH2*1 allele, compared to those with ADH2*2 allele[10,15], which were consistent to our findings. In contrast, Mizoi and his colleagues[24] did not find any significant differences of blood acetaldehyde levels among the three ADH2 genotype groups. However, in an in vitro study done by Yin and his colleagues[26], the enzyme activity in ADH2*2 allele was reported to be much higher that that of ADH2*1, suggesting that ethanol is much easily metabolized by ADH2*2 allele to become acetaldehyde. This finding conflicts with the observations from epidemiological studies[10,12,13,15,16] and our study as well. Thus, the detailed mechanism of this ADH2 polymorphism, which may be involved in another metabolic pathway of ethanol or closely linked with other functional polymorphisms, on esophageal cancer risk deserves further examination.
The frequency of ADH2 and ALDH2 genotypes varies across different racial groups (Table 5). ADH2*1 allele is found to be predominant in Caucasians (~90%)[27], whereas most of the Asian people carry the ADH2*2 allele[25,28]. In addition, the frequency of ALDH2*1 allele is higher in Caucasian populations than in Asian populations. These differences in gene distributions, which affect the metabolism of ethanol, may explain why the Asian people experience facial flushing and other marked adverse reactions to drinking alcoholic beverages more easily than Caucasians[29].
Our study found 20 case patients with the paired DNA specimens of tumors and peripheral blood to have exactly the same genotypes of ADH2 and ALDH2 between them. Although the sample size is relatively small, it can be speculated that ADH2 and ALDH2 genes might be involved in an early rather than an advanced stage of esophageal tumorigenesis. This is a hospital-based case-control study, but it is unlikely that individuals carrying one particular genotype were recruited differently in the cases and controls. In addition, in this study, the genotype distribution among the controls closely conformed to Hardy-Weinberg expected frequencies. Information about the consumption of alcoholic beverages was collected from questionnaires, which may randomly misclassify the interest of exposure between the cases and controls and result in the underestimation of our findings.
In conclusion, the present study found ADH2 and ALDH2 genotypes to be independently associated with esophageal cancer risk in the Taiwanese men. In addition, we have reported the interaction between the combined effect gene and environment on esophageal cancer risk. Subjects who consumed alcoholic beverages and carried susceptible genotypes of ADH2 and ALDH2 experienced a multiplicative increase in risk of developing esophageal cancer, much higher than those who were non-drinkers and did not carry susceptible genotypes of ADH2 and ALDH2. Our present findings can provide additional information about the role of alcohol on esophageal cancer risk in Taiwan.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Alcohol drinking. Biological data relevant to the evaluation of carcinogenic risk to humans. IARC Monogr Eval Carcinog Risks Hum. 1988;44:101-152. [PubMed] |
| 2. | Wu MT, Lee YC, Chen CJ, Yang PW, Lee CJ, Wu DC, Hsu HK, Ho CK, Kao EL, Lee JM. Risk of betel chewing for oesophageal cancer in Taiwan. Br J Cancer. 2001;85:658-660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 3. | Bosron WF, Li TK. Genetic polymorphism of human liver alcohol and aldehyde dehydrogenases, and their relationship to alcohol metabolism and alcoholism. Hepatology. 1986;6:502-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 391] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 4. | International Agency for Research on Cancer. Allyl compounds, aldehydes, epoxides and peroxides. IARC Monogr Eval Carcinoge Risk Chemi Hum. 1985;36:101-132. |
| 5. | Matsuo Y, Yokoyama R, Yokoyama S. The genes for human alcohol dehydrogenases beta 1 and beta 2 differ by only one nucleotide. Eur J Biochem. 1989;183:317-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Hsu LC, Bendel RE, Yoshida A. Direct detection of usual and atypical alleles on the human aldehyde dehydrogenase-2 (ALDH2) locus. Am J Hum Genet. 1987;41:996-1001. [PubMed] |
| 7. | Farrés J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994;269:13854-13860. [PubMed] |
| 8. | Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83:314-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 405] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 9. | Yokoyama A, Muramatsu T, Ohmori T, Higuchi S, Hayashida M, Ishii H. Esophageal cancer and aldehyde dehydrogenase-2 genotypes in Japanese males. Cancer Epidemiol Biomarkers Prev. 1996;5:99-102. [PubMed] |
| 10. | Hori H, Kawano T, Endo M, Yuasa Y. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and human esophageal squamous cell carcinoma susceptibility. J Clin Gastroenterol. 1997;25:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Yokoyama A, Muramatsu T, Ohmori T, Yokoyama T, Okuyama K, Takahashi H, Hasegawa Y, Higuchi S, Maruyama K, Shirakura K. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383-1387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 244] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Yokoyama A, Muramatsu T, Omori T, Matsushita S, Yoshimizu H, Higuchi S, Yokoyama T, Maruyama K, Ishii H. Alcohol and aldehyde dehydrogenase gene polymorphisms influence susceptibility to esophageal cancer in Japanese alcoholics. Alcohol Clin Exp Res. 1999;23:1705-1710. [RCA] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 59] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Yokoyama A, Muramatsu T, Omori T, Yokoyama T, Matsushita S, Higuchi S, Maruyama K, Ishii H. Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis. 2001;22:433-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 190] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Matsuo K, Hamajima N, Shinoda M, Hatooka S, Inoue M, Takezaki T, Tajima K. Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis. 2001;22:913-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Yokoyama A, Kato H, Yokoyama T, Tsujinaka T, Muto M, Omori T, Haneda T, Kumagai Y, Igaki H, Yokoyama M. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis. 2002;23:1851-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 169] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 16. | Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, Chu HC. Chinese alcoholic patients with esophageal cancer are genetically different from alcoholics with acute pancreatitis and liver cirrhosis. Am J Gastroenterol. 2000;95:2958-2964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Xu YL, Carr LG, Bosron WF, Li TK, Edenberg HJ. Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics. 1988;2:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 161] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry. 1996;168:762-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 119] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Harada S, Zhang S. New strategy for detection of ALDH2 mutant. Alcohol Alcohol Suppl. 1993;1A:11-13. [PubMed] |
| 20. | Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 1997;18:627-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 176] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Takeshita T, Kawai T, Morimoto K. Elevated levels of hemoglobin-associated acetaldehyde related to alcohol drinking in the atypical genotype of low Km aldehyde dehydrogenase. Cancer Res. 1997;57:1241-1243. [PubMed] |
| 22. | Yin SJ, Chou FJ, Chao SF, Tsai SF, Liao CS, Wang SL, Wu CW, Lee SC. Alcohol and aldehyde dehydrogenases in human esophagus: comparison with the stomach enzyme activities. Alcohol Clin Exp Res. 1993;17:376-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 91] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Enomoto N, Takase S, Yasuhara M, Takada A. Acetaldehyde metabolism in different aldehyde dehydrogenase-2 genotypes. Alcohol Clin Exp Res. 1991;15:141-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 190] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol. 1994;29:707-710. [PubMed] |
| 25. | Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677-681. [PubMed] |
| 26. | Yin SJ, Bosron WF, Magnes LJ, Li TK. Human liver alcohol dehydrogenase: purification and kinetic characterization of the beta 2 beta 2, beta 2 beta 1, alpha beta 2, and beta 2 gamma 1 "Oriental" isoenzymes. Biochemistry. 1984;23:5847-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 115] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, Bhatia K, Chen LZ, Fang B, Lisker R. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 373] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 28. | Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152:1219-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 206] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 29. | Wolff PH. Ethnic differences in alcohol sensitivity. Science. 1972;175:449-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 224] [Article Influence: 4.2] [Reference Citation Analysis (0)] |