Published online Aug 7, 2005. doi: 10.3748/wjg.v11.i29.4587
Revised: January 7, 2005
Accepted: January 14, 2005
Published online: August 7, 2005
AIM: To study the expressions of p27kip1 protein and p27mRNA, the hypermethylation of p27kip1 and the relation between them in various stages of hepatocarcinogenesis.
METHODS: p27 protein and p27mRNA were detected by immunohistochemical staining and in situ hybridization respectively in 68 cases of normal liver, liver cirrhosis, pericancerous cirrhosis and hepatocellular carcinoma (HCC). The hypermethylation of p27kip1 was detected by methylation-specific PCR (MSP) in 44 cases of normal liver, liver cirrhosis, and HCC.
RESULTS: The positive rate of p27 protein was 66.7% (4/6) in normal liver, 60.0% (6/10) in liver cirrhosis, 50.0% (12/24) in pericancerous cirrhosis and 21.4% (6/28) in HCC. There were no statistical differences in normal liver, liver cirrhosis and pericancerous cirrhosis, but the positive rate of p27 protein significantly decreased in HCC compared to that in the other groups (P = 0.006, χ2 = 7.664). The positive rate of p27kip1 mRNA was 83.3% (5/6) in normal liver, 70.0% (7/10) in liver cirrhosis, 75.0% (18/24) in pericancerous cirrhosis and 25.0% (7/28) in HCC. There were no statistical differences in normal liver, liver cirrhosis and pericancerous cirrhosis, but the positive rate of p27kip1 mRNA also significantly decreased in HCC compared to that in the other groups (P = 0.000, χ2 = 16.600). In addition, there was a significant correlation between the expression of p27 protein and p27mRNA in the integrated group of normal liver and liver cirrhosis. However, no significant correlation was found between pericancerous cirrhosis and HCC. Using MSP, we found that 1 HCC in 44 cases (including 6 cases of normal liver, 10 cases of liver cirrhosis and 28 cases of HCC) was methylated, whose p27 protein and p27mRNA were negative.
CONCLUSION: The reduction or loss of p27 protein and p27mRNA are potentially involved in hepatocarcinogenesis. The hypermethylation of p27 might lead to the loss of p27mRNA transcription.
- Citation: Lei PP, Zhang ZJ, Shen LJ, Li JY, Zou Q, Zhang HX. Expression and hypermethylation of p27kip1 in hepatocarcinogenesis. World J Gastroenterol 2005; 11(29): 4587-4591
- URL: https://www.wjgnet.com/1007-9327/full/v11/i29/4587.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i29.4587
Disruption of cell cycle regulation plays a significant role in carcinogenesis. p27kip1, located at chromosome 12p13, is a regulator of the mammalian cell cycle and a tumor suppressor[1]. It was reported that the expression of p27kip1 protein in hepatocellular carcinoma (HCC) is lost or reduced, when compared to that in liver cirrhosis and normal liver[2,3] as well as the transcription of p27mRNA[4]. The mechanism of loss and reduction of p27kip1 expression during hepatocarcinogenesis still remains unclear. Specific alterations of the p27kip1gene, including mutations and homozygous deletions, are exceedingly rare in HCC, suggesting that p27kip1 might be inactivated by transcription rather than genomic aberrations.
In normal tissues, methylation of the promoter region CpG islands is associated with transcriptional silencing of imprinted alleles and genes on the inactive X chromosome[5,6]. Indeed, aberrant DNA methylation of the promoter region CpG islands can serve as an alternative to mutations in the coding region for the inactivation of tumor suppressor genes, including the APC gene, p16INK4A, and p15INK4B[7-9]. p27kip1 promoter methylation has not been studied in HCC. We focused on analyzing the DNA methylation patterns in CpG islands of the p27kip1 gene.
Specimens obtained from surgical resection, autopsy of livers from 1970 to 2003 were fixed in 40 g/L formaldehyde, embedded in paraffin, and stained with routine HE. The specimens were divided into four groups: normal liver tissue specimens used as controls (n = 6), liver cirrhosis tissue specimens (n = 10), pericancerous tissue specimens (n = 24) and HCC tissue specimens (n = 28). All specimens were examined by two pathologists.
Immunohistochemistry S-P method was used to detect p27kip1 protein expression. Mouse monoclonal antibody to human p27kip1 was purchased from Fuzhou Maixin Biotechnical Company. The main steps were as follows. The tissues were treated with 3% H2O2 to block endogenous peroxidase at room temperature for 10 min, heated to boiling for 5 min in 10 mmol/L sodium citrate (pH 6.0) buffer in a pressure cooker, incubated in endogenous peroxidase blocking solution at room temperature for 10 min, and then incubated in normal nonimmune serum at room temperature for 10 min. The mouse anti p27kip1 antibody was added to tissue sections and incubated overnight at 4 °C. Biotin-conjugated secondary antibody was added to the sections and incubated at room temperature for 10 min. S-P complex was added at room temperature for 10 min and then DAB was used for the color reaction. The tissue sections were washed with PBS (0.01 mol/L, pH 7.4) between each step. Positive and negative controls were simultaneously used to ensure specificity and reliability of the staining process. A positive section provided by the company was taken as positive control. In the negative control, PBS was used to replace the primary antibody.
The immunohistochemical staining (IHC) was independently assessed by two experienced pathologists in a double-blind fashion. Positive p27 staining was mainly localized in the cell nuclei and only rarely in the cytoplasm. All fields of each section were observed. If there was no positive cell, the grade was 0; under 30% of positive cells the grade was 1; 31-70% positive cells the grade was 2; >70% positive cells the grade was 3. The criterion of the staining intensity was determined by the staining characteristics of most cells in each section. If there was no staining, the scale was 0; weak yellow staining was grade 1; brown yellow staining was grade 2; brown staining was grade 3. The final staining results were determined by the total of the staining and intensity grade. If the total grade was 0, the result was regarded as negative (-); grades between 1 and 3 as weakly positive (+); grades between 4 and 6 as strongly positive (++)[10].
In situ hybridization (ISH) was used for the detection of p27kip1 mRNA. The p27kip1 probe with digoxin-labeled and ISH kit was purchased from Wuhan Boster Biological Technology Ltd. The main steps were as follows. All slides were baked overnight at 58-60 °C. The tissues were deparaffinized by xylene and graded alcohols, treated with 3% H2O2 at room temperature for 10 min. Proteinase K freshly diluted with 3% citrate acid was added at 37 °C for 10-15 min. The tissues were pre-hybridized for 3 h at 37 °C in pre-hybridization liquid. Digoxin-labeled probe with coverslip was added, the tissues were hybridized at 37 °C for about 16 h. The slides were washed four times with SSC at 37 °C. Block liquid was added for 3 min at 37 °C. Mouse biotin-antidigoxin antibody was applied for 60 min at 37 °C, followed by detection with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) for 20 min. Positive control provided by the company was used. Hybridization liquid replaced by PBS served as negative control.
ISH staining was evaluated according to the following scales: no positive cell for negative, 1+ for staining in <25% of cells, 2+ for staining in 25-70% of cells, and 3+ for staining in >70% of cells[11].
Tissues were deparaffinized by xylene and graded alcohols. Genomic DNA was then extracted with a standard phenol/chloroform procedure. DNA methylation patterns in the CpG islands of the p27kip1 gene were determined by methylation-specific PCR (MSP)[12]. MSP distinguishes unmethylated from methylated alleles based on sequence changes produced after bisulfite treatment of DNA, which converts unmethylated (but not methylated) cytosine to uracil, and subsequent PCR using primers were designed for either methylated or unmethylated DNA. Sodium bisulfite modification was performed using the CpGenomeTM DNA modification kit (Intergen, Oxford, UK) according to the manufacturer’s protocol with minor modifications. Briefly, DNA was denatured by NaOH (final concentration, 0.2 mol/L) for 15 min at 37 °C. Sodium bisulfite solution at pH 5.0, freshly prepared, was added (550 µL), and incubated at 50 °C for 20 h. The modified DNA was treated with NaOH (final concentration, 0.3 mol/L) for 5 min at room temperature. After precipitation by ethanol, the DNA was resuspended in TE buffer (10 mmol/L Tris pH 8.0, 0.1 mmol/L EDTA)[13]. The primer sequences for the p27kip1 methylated reaction were 5’-AAGAGGCGAG-TTAGCGT-3’ (sense) and 5’-AAAACGCCGCCGAACGA-3’ (antisense); 5’-ATGGAAGAGGTGAGTTAGT-3’ (sense) and 5’-AAAACCCCAATTAAAAACA-3’ (antisense) for the p27kip1 unmethylated reaction[14]. PCR was carried out in a 10 µL volume containing PCR buffer (10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2), dNTPs (250 µmol/L each), primers (2.5 pmoL each), 0.4 unit of TaKaRa taqTM (DR100A, TaKaRa Biotech, Dalian, China) and approximately 10-100 ng bisulfite-modified DNA. Amplification was carried out in a DNA Autorisierter thermocycler (Eppendorf) with initial denaturing at 95 °C for 3 min followed by 34 cycles of denaturing at 94 °C for 1 min, annealing for 1 min at 53 °C (for p27kip1 methylated reaction) or at 58 °C (for p27kip1 unmethylated reaction), extension for 1 min at 72 °C, and then a final extension for 10 min at 72 °C. DNA extracted from HCC treated with methylase (SssI, New England BioLabs, Beverly, MA, USA) was used as the methylated control and DNA extracted from relatively normal liver was the unmethylated control. Amplified products were electrophoresed on 2% agarose gels and visualized with ethidium bromide staining. In the total, we tested 44 cases including 6 cases of normal liver, 10 cases of liver cirrhosis and 28 cases of HCC.
The Fisher’s exact test, χ2 test and Spearman’s correlation were used.
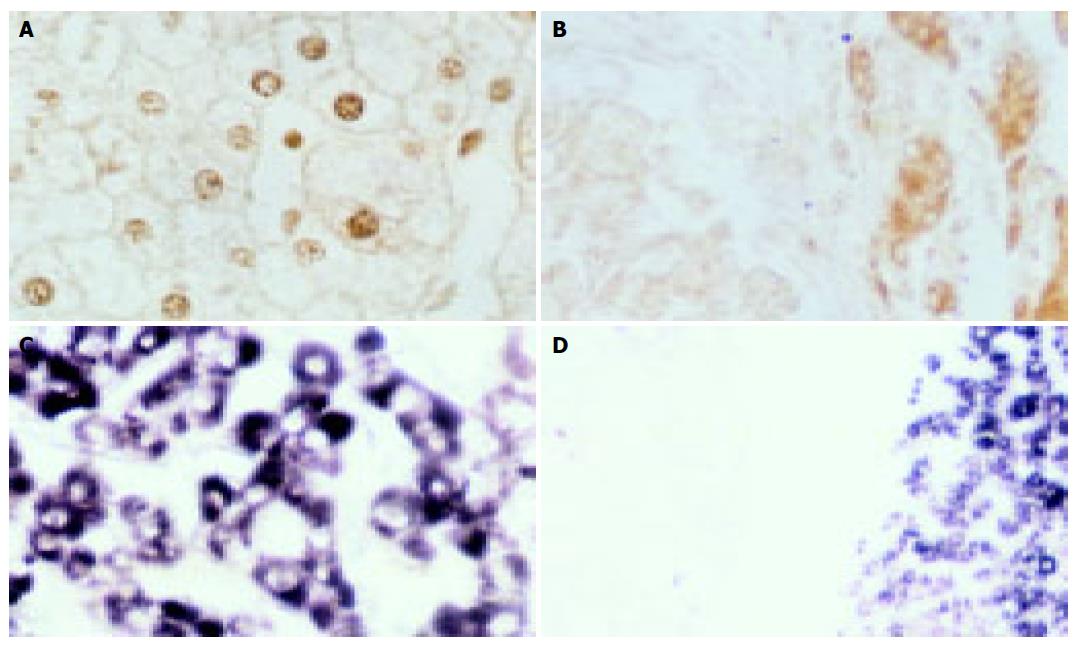
The positive rate of p27 protein was 66.7% (4/6) in normal liver, 60.0% (6/10) in liver cirrhosis, 50.0% (12/24) in pericancerous cirrhosis and 21.4% (6/28) in HCC. There were no statistical differences in normal liver, liver cirrhosis and pericancerous cirrhosis, but the positive rate of p27 protein significantly decreased in HCC compared to that in the other groups (P = 0.006, χ2 = 7.664). In addition, the positive signals of p27 protein were mainly located in nuclei in normal liver and liver cirrhosis (Figure 1A), while they were located in cytoplasm in pericancerous cirrhosis and HCC (Figure 1B).
The positive result showed blue coloration in the cytoplasm (Figures 1C and D). The positive rate of p27kip1 mRNA was 83.3% (5/6) in normal liver, 70.0% (7/10) in liver cirrhosis, 75.0% (18/24) in pericancerous cirrhosis and 25.0% (7/28) in HCC. There were no statistical differences in normal liver, liver cirrhosis and pericancerous cirrhosis, but the positive rate of p27kip1 mRNA also significantly decreased in HCC compared to that in the other groups (P = 0.000, χ2 = 16.600). There was a significant correlation between the expression of p27 protein and p27mRNA in the integrated group of normal liver and liver cirrhosis (P = 0.082<0.1, χ2 = 0.447, r = 0.447, Table 1). However, no significant correlations were between pericancerous cirrhosis (P = 0.368, χ2 = 0.192, r = 0.192) and HCC (P = 0.611, χ2 = 0.101, r = 0.101, Tables 2 and 3).
| p27 protein | p27mRNA | Total | |
| + | – | ||
| + | 9 | 1 | 10 |
| – | 3 | 3 | 6 |
| Total | 12 | 4 | 16 |
| p27 protein | p27mRNA | Total | |
| + | – | ||
| + | 10 | 2 | 12 |
| – | 8 | 4 | 12 |
| Total | 18 | 6 | 24 |
| p27 protein | p27mRNA | Total | |
| + | – | ||
| + | 2 | 4 | 6 |
| – | 5 | 17 | 22 |
| Total | 7 | 21 | 28 |
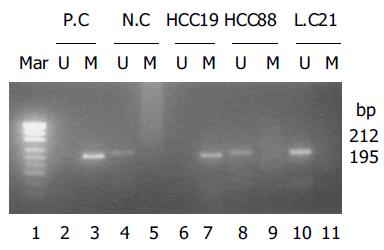
p27kip1 promoter hypermethylation was only detected in 1 of 28 HCC cases, which was negative when detected by ISH and IHC. p27kip1 methylation was not detected in any of 10 cases of liver cirrhosis and 6 cases of normal livers. Methylated and unmethylated control DNA showed the expected fragment sizes of 195 and 212 bp (Figure 2).
p27 is involved in cancer because of its essential role in controlling cell cycle progression[1]. Decreased expression of p27 protein is inversely correlated with stage of disease at the time of surgery in HCC, reduced p27 is significantly related to advanced locoregional extent of the primary tumor and tumor size[15]. The expression of p27 is a favorable prognostic indicator in patients with HCC[16] and other tumors[17,18]. Our results revealed that p27 protein was expressed in normal liver, liver cirrhosis, pericancerous cirrhosis and HCC, but the positive expression rate of p27 protein significantly decreased in HCC compared to that in the other groups. The loss or decrease of p27 protein may lead to reduction or disappearance of its cell cycle negative regulation, thus cells pass the G1 phase into S phase, resulting in division and autonomous program[19].
Our results also revealed that the positive rate of p27mRNA was high in the normal liver, liver cirrhosis and pericancerous cirrhosis, and there were no differences in them. But the level of p27mRNA was low in HCC compared to that in the other groups, suggesting that reduced p27 expression may be at least partly responsible for human hepatocarcinogenesis[4]. There was a significant correlation between the expression of p27 protein and p27mRNA in the integrated group of normal liver and liver cirrhosis. However, no significant correlations were found between pericancerous cirrhosis and HCC. This phenomenon has not been reported. Whether the expression of p27 protein is regulated by p27mRNA before carcinogenesis or by other mechanisms remains to be verified.
After having analyzed 44 cases (including 6 cases of normal liver, 10 cases of liver cirrhosis and 28 cases of HCC) by MSP, we found that one HCC case was hypermethylated, whose p27 protein and p27mRNA were negative, suggesting that p27 methylation might lead to its loss of transcription, contributing to cell proliferation. The same result has been reported in a few other tumors[20-22], but not in HCC[23]. Because there are few reports on the p27 methylation in HCC, only one HCC case was hypermethylated in our study, we could not analyze the relationship between the p27mRNA expression and p27 gene methylation.
In conclusion, reduction or loss of p27 protein and p27mRNA is potentially involved in hepatocarcinogenesis, and hypermethylation of p27 may lead to the loss of p27mRNA transcription[20-22].
Co-first-authors: Pu-Ping Lei and Zong-Ji Zhang
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massagué J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 1611] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 2. | Ito Y, Matsuura N, Sakon M, Miyoshi E, Noda K, Takeda T, Umeshita K, Nagano H, Nakamori S, Dono K. Expression and prognostic roles of the G1-S modulators in hepatocellular carcinoma: p27 independently predicts the recurrence. Hepatology. 1999;30:90-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 120] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 3. | Chen TC, Ng KF, Lien JM, Jeng LB, Chen MF, Hsieh LL. Mutational analysis of the p27(kip1) gene in hepatocellular carcinoma. Cancer Lett. 2000;153:169-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Hui AM, Sun L, Kanai Y, Sakamoto M, Hirohashi S. Reduced p27Kip1 expression in hepatocellular carcinomas. Cancer Lett. 1998;132:67-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol. 2001;195:97-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 184] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 6. | Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157-3173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 241] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Tsujie M, Yamamoto H, Tomita N, Sugita Y, Ohue M, Sakita I, Tamaki Y, Sekimoto M, Doki Y, Inoue M. Expression of tumor suppressor gene p16(INK4) products in primary gastric cancer. Oncology. 2000;58:126-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H. Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest. 2001;81:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 109] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K. DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn J Cancer Res. 2002;93:767-773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Zhao JG, Wu AG, Huang ZH, Yang JH. Expression of p27 and its clinical significance in colorectal carcinomas. Chin Ger J Clin Oncol. 2002;l:129-131. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 11. | Erickson LA, Jin L, Wollan P, Thompson GB, van Heerden JA, Lloyd RV. Parathyroid hyperplasia, adenomas, and carcinomas: differential expression of p27Kip1 protein. Am J Surg Pathol. 1999;23:288-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 72] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4248] [Article Influence: 146.5] [Reference Citation Analysis (0)] |
| 13. | Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, Kleihues P, Ohgaki H. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 151] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Nakamura M, Sakaki T, Hashimoto H, Nakase H, Ishida E, Shimada K, Konishi N. Frequent alterations of the p14(ARF) and p16(INK4a) genes in primary central nervous system lymphomas. Cancer Res. 2001;61:6335-6339. [PubMed] |
| 15. | Tannapfel A, Grund D, Katalinic A, Uhlmann D, Köckerling F, Haugwitz U, Wasner M, Hauss J, Engeland K, Wittekind C. Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Fiorentino M, Altimari A, D'Errico A, Cukor B, Barozzi C, Loda M, Grigioni WF. Acquired expression of p27 is a favorable prognostic indicator in patients with hepatocellular carcinoma. Clin Cancer Res. 2000;6:3966-3972. [PubMed] |
| 17. | Nitti D, Belluco C, Mammano E, Marchet A, Ambrosi A, Mencarelli R, Segato P, Lise M. Low level of p27(Kip1) protein expression in gastric adenocarcinoma is associated with disease progression and poor outcome. J Surg Oncol. 2002;81:167-75; discussion 175-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Anastasiadis AG, Calvo-Sanchez D, Franke KH, Ebert T, Heydthausen M, Schulz WA, Burchardt M, Gerharz CD. p27KIP1-expression in human renal cell cancers: implications for clinical outcome. Anticancer Res. 2003;23:217-221. [PubMed] |
| 19. | Filipits M, Puhalla H, Wrba F. Low p27Kip1 expression is an independent prognostic factor in gallbladder carcinoma. Anticancer Res. 2003;23:675-679. [PubMed] |
| 20. | Qian X, Jin L, Kulig E, Lloyd RV. DNA methylation regulates p27kip1 expression in rodent pituitary cell lines. Am J Pathol. 1998;153:1475-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Worm J, Bartkova J, Kirkin AF, Straten P, Zeuthen J, Bartek J, Guldberg P. Aberrant p27Kip1 promoter methylation in malignant melanoma. Oncogene. 2000;19:5111-5115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Nakatsuka S, Liu A, Yao M, Takakuwa T, Tomita Y, Hoshida Y, Nishiu M, Aozasa K. Methylation of promoter region in p27 gene plays a role in the development of lymphoid malignancies. Int J Oncol. 2003;22:561-568. [PubMed] |
| 23. | Yu J, Zhang HY, Ma ZZ, Lu W, Wang YF, Zhu JD. Methylation profiling of twenty four genes and the concordant methylation behaviours of nineteen genes that may contribute to hepatocellular carcinogenesis. Cell Res. 2003;13:319-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |