Published online Jul 21, 2005. doi: 10.3748/wjg.v11.i27.4180
Revised: November 14, 2004
Accepted: November 19, 2004
Published online: July 21, 2005
AIM: To establish a simplified method for generating peptide-major histocompatibility complex (MHC) class I tetramers.
METHODS: cDNAs encoding the extracellular domain of human lymphocyte antigen (HLA)-A*0201 heavy chain (A2) and β2-microglobulin (β2m) from total RNA extracted from leukocytes of HLA-A2+ donors were cloned into separate expression vectors by reverse transcription-polymerase chain reaction. The recombinant A2 and β2m proteins were expressed in Escherichia coli strain BL21(DE3) and recovered from the inclusion body fraction. Soluble A2 proteins loaded with specific antigen peptides were refolded by dilution from the heavy chain in the presence of light chain β2m and HLA-A2-restricted peptide antigens. The refolded A2 monomers were biotinylated with a commercial biotinylation enzyme (BirA) and purified by low pressure anion exchange chromatography on a Q-Sepharose (fast flow) column. The tetramers were then formed by mixing A2 monomers with streptavidin-PE in a molar ratio of 4:1. Flow cytometry was used to confirm the expected tetramer staining of CD8+ T cells.
RESULTS: Recombinant genes for HLA-A*0201 heavy chain (A2) fused to a BirA substrate peptide (A2-BSP) and mature β2m from HLA-A2+ donor leukocytes were successfully cloned and highly expressed in E. coli. Two soluble monomeric A2-peptide complexes were reconstituted from A2-BSP in the presence of β2m and peptides loaded with either human cytomegalovirus pp65495-503 peptide (NLVPMVATV, NLV; designated as A2-NLV) or influenza virus matrix protein Mp58–66 peptide (GILGFVFTL, GIL; designated as A2-GIL). Refolded A2-NLV or A2-GIL monomers were biotinylated and highly purified by single step anion exchange column chromatography. The tetramers were then formed by mixing the biotinylated A2-NLV or A2-GIL monomers with streptavidin-PE, leading to more than 80% multiplication as revealed by SDS-PAGE under non-reducing, unboiled conditions. Flow cytometry revealed that these tetramers could specifically bind to CD8+ T cells from a HLA-A2+ donor, but failed to bind to those from a HLA-A2- donor.
CONCLUSION: The procedure is simple and efficient for generating peptide-MHC tetramers.
- Citation: He XH, Xu LH, Liu Y. Procedure for preparing peptide-major histocompatibility complex tetramers for direct quantification of antigen-specific cytotoxic T lymphocytes. World J Gastroenterol 2005; 11(27): 4180-4187
- URL: https://www.wjgnet.com/1007-9327/full/v11/i27/4180.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i27.4180
Cytotoxic T lymphocytes (CTLs), or CD8+ T cells, play a critical role in the clearance of viral infections and the eradication of tumors[1]. A better understanding of cellular immunity against viruses and/or tumor cells requires careful analysis of the responding CD8+ T cells, particularly in terms of their numbers and effector functions[2]. Because specific CD8+ T cells are often vastly outnumbered by irrelevant T cells, quantification of antigen-specific T cells often requires in vitro culture and re-stimulation, which may introduce bias in the results[2]. The standard method for deriving specific CTL frequency information is limiting dilution analysis (LDA)[2]. However, accumulating evidence indicates that this technique may significantly underestimate the number of specific CTLs, because it does not detect cells that lack proliferative potential[2,3]. This limitation has been overcome by the introduction of peptide-major histocompatibility complex (MHC) class I tetrameric complex technology[3-6], which initiates a profound revolution in the field of cellular immunology[7-10]. Peptide-MHC tetramer-based assays have enabled direct flow cytometric quantification[3-6], phenotyping[3,8-11] and even functional analysis[8,11] of antigen-specific CD8+ T cells. Thus, peptide-MHC tetramer technology is a powerful tool for evaluating the fundamental aspects of T-cell immunity.
Though tetramer technology for direct quantification of the frequency and/or function of antigen-specific CTLs has been generally accepted, wide application of tetramer strategies is limited by the complex procedures necessary for tetramer production[3]. Moreover, the expression level of recombinant human MHC class I heavy chain in Escherichia coli is usually minimal due to the low translation frequency of human proteins in E. coli[12]. To address these problems, we sought to develop a simplified and efficient method for production of peptide-MHC class I tetramers. We constructed prokaryotic expression vectors for recombinant human lymphocyte antigen (HLA)-A2 heavy chain and β2-microglobulin (β2m) proteins, in which nucleotide residues in the translation initiation region (TIR) were substituted to the preferred codons for E. coli[13] and decreased the G/C content in this region[14]. The recombinant A2 and β2m proteins were overexpressed in E. coli in the form of inclusion bodies, thus facilitating their high purity isolation by simple washing and centrifugation. Further simple procedures allowed us to establish an efficient, streamlined procedure for the preparation of HLA-A2 tetramers. This improved procedure should facilitate the general application of tetramer technology in both basic research and clinical applications.
E. coli strain DH5α was stored in our laboratory. E. coli strain BL21(DE3) and plasmid pET-3c were purchased from Novagen (Madison, WI, USA). NdeI, BamHI, T4 DNA ligase and high fidelity DeepVent Taq polymerase were purchased from New England Biolabs (Beverly, MA, USA). The TRIzol reagent and ThermoScript reverse transcription-polymerase chain reaction (RT-PCR) system were obtained from Invitrogen (Carlsbad, CA, USA). Q-Sepharose (fast flow) was obtained from Amersham (Uppsala, Sweden). Mouse anti-human monoclonal antibodies CD3-FITC, CD8-CyChrome and HLA-A2-FITC were purchased from PharMingen (San Diego, CA, USA). Streptavidin R-phycoerythrin (PE) conjugate (streptavidin-PE) was purchased from Molecular Probes (Eugene, OR, USA). Protein molecular mass markers were obtained from Sigma (St. Louis, MO, USA). The biotinylation enzyme, BirA, was purchased from Avidity (http://www.avidity.com). The NLVPMVATV (NLV) peptide derived from pp65 (pp65495-503) of HCMV[11], and the GILGFVFTL (GIL) peptide derived from the matrix protein (Mp[58-66]) of the influenza A virus[15], were synthesized by BioAsia (Shanghai, China) and purified to >98%. All other chemicals were from Sigma and were analytically pure.
Heparinized human peripheral blood was collected from three HLA-A2 positive (identified by anti-human HLA-A2-FITC staining and flow cytometric analysis) donors by venipuncture. Total RNA was extracted from freshly isolated PBMCs using the TRIzol reagent. cDNAs were synthesized from the isolated RNA using the ThermoScript RT-PCR system according to the recommended procedure. PCR amplification of the resultant cDNA was performed in a total volume of 50 μL containing high fidelity DeepVent Taq polymerase. For amplification of β2m, PCR was performed with an initial denaturation for 2 min at 94°C, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, and a final extension for 10 min at 72°C. The primers (5’-ATA TCC ATA TGT CTC GCT CCG TGG CCT TAG-3’ and 5’-AAC TAG GGA TCC TTA CAT GTC TCG ATC CCA C-3’) were designed to amplify the entire coding sequence of human β2m cDNA. For amplification of the mature HLA-A*0201 heavy chain, PCR was performed as above except that the extension time at 72°C was 1.5 min and the primers (5’-TAT ACA TAT GGG CTC TCA CTC CAT GAG GTA TTT C-3’ and 5’-AAC CAG GGA TCC TAC ACT TTA CAA GCT GTG AGA G-3’) were designed to amplify the entire coding sequence of mature A2. The resultant PCR products were cloned into the NdeI–BamHI sites of the pET-3c vector. Randomly selected clones were NdeI/BamHI digested and screened for the presence of a correctly sized insert. Several independent clones were submitted for DNA sequencing of the HLA-A*0201 heavy chain cDNA using the dye-labeled deoxy-terminator protocol on a 377 automated DNA sequencer (Applied Biosystems). The cloned β2m cDNA was also confirmed by DNA sequencing analysis.
To create a β2m expression construction with optimized codon usage and G/C content, the DNA fragment for mature β2m was PCR amplified from the cloned β2m cDNA using specific primers (5’-AT ATC CAT ATG ATT CAA CGT ACT CCA AAA ATT CAA GTT TAC TCA CGT CAT CC-3’ and 5’-CGA CTG GAT CCT TAC ATG TCT CGA TCC CAC TTA AC-3’). The underlined codons were optimized for expression in E. coli[13] by synonymous substitutions from ATC, CAG, AAG, CAG to ATT, CAA, AAA and CAA, respectively. These alterations also reduced the G/C content in the TIR. The resulting PCR product was inserted into the NdeI-BamHI sites of the pET-3c vector, and a positive clone with a correct sized insert was confirmed by DNA sequencing. This recombinant plasmid was designated as pET-β2m.
To construct an expression vector in which the HLA-A2 heavy chain was fused with BSP, the DNA fragment encoding a Gly-Ser linker and a BSP (LHHILDAQKMVWNHR) was fused to the 3’ end of the cDNA encoding the extracellular domain of the HLA-A2 heavy chain (1-275) by PCR amplification from cloned HLA-A2 heavy chain cDNA with specific primers (5’- ATA CAT ATG GGT TCT CAT TCT ATG CGT TAT TTT TTT ACA TCT GTT TCC CGG CCC GGC CGC-3’ and the 3’ primer 5’-GCG CGG ATC CTT AAC GAT GAT TCC ACA CCA TTT TCT GTG CAT CCA GAA TAT GAT GCA GAG AGC CCG GCT CCC ATC TCA GGG T-3’). The underlined codons were optimized for expression in E. coli[13], and reduced the G/C content without any changes in amino acid sequence. These codons were changed from GGC, CAC, TCC, AGG, TTC, TTC, TCC and GTG, respectively. The resultant PCR product was NdeI/BamHI digested and subcloned into plasmid pET-3c. Clones with correct sized inserts were verified by direct sequencing, and the recombinant plasmid was designated as pET-A2-BSP.
BL21(DE3) competent cells were transformed with either pET-β2m or pET-A2-BSP, single colonies were used to inoculate 100 mL of LB medium, and cultures were incubated at 37°C overnight until cells reached the stationary phase. Each stationary culture was diluted 10-fold with fresh LB medium (to 1 L) and incubated at 37°C for a further 2 h, 0.4 mmol/L isopropyl β-D-thiogalactopyranoside (IPTG) was added, and cells were incubated for an additional 4 h. Cells were collected by centrifugation, and insoluble protein aggregates (inclusion bodies) were purified essentially as described[16]. In brief, each cell pellet was re-suspended in 50 mmol/L Tris-HCl buffer (pH 8.0) containing 1 mmol/L EDTA, 0.1 mmol/L phenylmethylsulfonyl fluoride (PMSF) and 10 mmol/L dithiothreitol (DTT), and then sonicated on ice. The insoluble pellet was collected by centrifugation and washed with washing buffer (50 mmol/L Tris-HCl pH 8.0, 100 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L DTT and 5 g/L Triton X-100), followed by three more rounds of sonication, centrifugation and collection. The pellet was washed three more times with washing buffer without Triton X-100 and the isolated inclusion body pellet was dissolved in 20 mmol/L 2-(N-morpholino)ethanesulfonic acid (pH 6.0, containing 8 mol/L urea, 10 mmol/L EDTA, and 0.1 mmol/L DTT). The insoluble material was pelleted by centrifugation and removed. The protein concentration of the remaining solution was determined by measuring A280 nm and A260 nm, and calculated according to the empirical formula (1.45×A280 nm-0.74×A260 nm = protein concentration in mg/mL). The protein solution was immediately frozen at -70°C.
The HLA-A2 monomers were refolded essentially as described by Garboczi and Wiley[17], with slight modifications. Briefly, 2 mg of peptide (NLV or GIL peptide) dissolved in DMSO was added in drops to 200 mL of pre-chilled refolding buffer (100 mmol/L Tris-HCl pH 8.0, containing 400 mmol/L L-arginine, 2 mmol/L EDTA, 5 mmol/L reduced glutathione, 0.5 mmol/L oxidized glutathione and 0.2 mmol/L PMSF). Then, 6 mg A2-BSP in 1 mL injection buffer containing 3 mol/L guanidine–HCl pH 4.2, 10 mmol/L sodium acetate and 10 mmol/L EDTA was forcefully injected to the stirring reaction through a 26-gauge needle as close to the stir bar as possible. Five micrograms of β2m was injected similarly, and the refolding mixture was incubated at 10°C for 3 d. At the end of the incubation, 200 mL of the refolding mixture was concentrated to 5 mL with an ultrafiltration apparatus (Amicon, Millipore, Bedford, MA, USA) containing a 10 ku molecular mass cut-off membrane, and dialyzed against 10 mmol/L Tris-HCl buffer (pH 8.0, containing 0.2 mmol/L PMSF). The refolded monomeric HLA-A2 was centrifuged to eliminate precipitates, and then biotinylated.
The refolded HLA-A2 was enzymatically biotinylated by incubation with BirA according to the procedure provided by the supplier (Avidity Co.). The biotinylated HLA-A2 was dialyzed against 10 mmol/L Tris-HCl buffer (pH 8.0, containing 0.2 mmol/L PMSF) and loaded onto an anion-exchanger Q-Sepharose column (2 cm×8 cm) equilibrated with the same buffer. The column was eluted with a 0-300 mmol/L NaCl linear gradient, and 1.5 mL fraction was collected. Fractions exhibiting both HLA-A2 heavy chain and β2m bands upon SDS-PAGE analysis were pooled and concentrated to 300 µL. The buffer of the biotinylated HLA-A2 was changed to 0.01 mol/L phosphate-buffered saline (PBS, pH 7.4) containing 0.2 mmol/L PMSF and 2 mmol/L EDTA through ultrafiltration.
HLA-A2 tetramers were formed by mixing the biotinylated proteins with streptavidin-PE at a molar ratio of 4:1. Tetramers were analyzed by SDS-PAGE of samples prepared without boiling, in a loading buffer that contained no reducing reagent[10].
Peripheral blood mononuclear cells (PBMCs) were separated from 3 mL whole blood of two volunteers (over 40 years) by centrifugation over a Ficoll-Hypaque density gradient (Lymphoprep; NYCOMED, Norway). For staining, 1×106 cells suspended in 50 µL PBS containing 20 mL/L fetal calf serum and 1 g/L sodium azide were incubated on ice for 1 h with 10 µL of CD3-FITC, 10 µL of CD8-CyChrome and 1 µg of HLA-A2 tetramer. After being washed, the cells were fixed in 300 µL of 10 g/L paraformaldehyde and detected on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). For each sample, 50 000 events were collected and analyzed with the CELLQuest software (Becton Dickinson). The results were expressed as the percentages of tetramer-binding cells in the total T cell population.
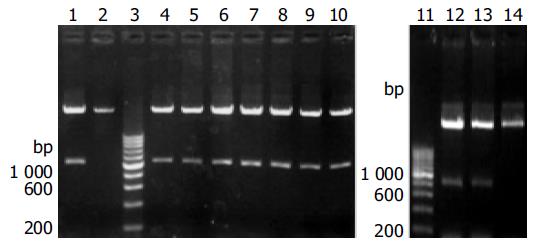
The cDNA encoding the mature HLA-A2 heavy chain was RT-PCR amplified from total RNA of three HLA-A2 positive donors (identified as such by flow cytometry after anti-HLA-A2-FITC staining). The DNA fragments with the expected length (1 100 bp) were inserted into pET-3c, and eight independently transformed DH5a clones were identified to have the correct insert (Figure 1). DNA sequencing of these clones showed that six clones from donors 1 and 2 contained the cDNA for HLA-A*0201 heavy chain (designated pET-A2), while those from donor 3 contained the cDNA for HLA-A*0207 heavy chain. The cDNA sequence for HLA-A*0201 was submitted to GenBank (accession number AY191309).
Similarly, the cDNA encoding β2m was cloned from the total RNA of one donor and inserted into pET-3c (data not shown). The sequence was verified by DNA sequencing and found to be identical to the published one (submitted to GenBank, accession number AY187687).
According to Altman’s strategy[3], the DNA fragment encoding a Gly-Ser linker and a BSP (LHHILDAQKMVWNHR) was fused to the 3’ end of the DNA fragment encoding the extracellular domain of HLA-A*0201 heavy chain (residues from 1 to 275) by PCR amplification of plasmid pET-A2. In order to increase the expression level in E. coli, our primers introduced synonymous substitutions at the 5’ region, intended to reduce the G/C content of the TIR and to optimize codons for the bias usage of E. coli[13,14]. The amplified DNA fragment (900 bp) was inserted into pET-3c. Two clones with the correct insert were confirmed by DNA sequencing, and the generated expression vector was designated as pET-A2-BSP (Figure 1).
The expression vector for mature β2m was similarly constructed by PCR amplification using cloned β2m cDNA as template. The codons at the 5’ region were optimized for better expression of β2m in E. coli. The sequence of the insert was verified by DNA sequencing, and the vector was designated as pET-β2m.
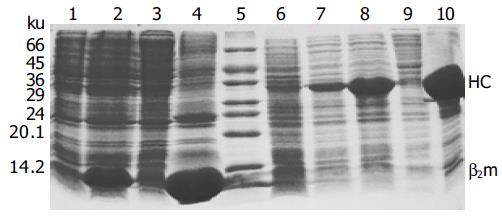
The expression vectors pET-A2-BSP and pET-β2m were transformed into E. coli BL21(DE3). The cells showed leaky expression of the two proteins, and the expression levels increased dramatically after induction with IPTG (Figure 2). The molecular mass of the recombinant β2m was approximately 12 ku, which was consistent with that of native human β2m, while the engineered A2 heavy chain-BSP fusion protein (A2-BSP) had the expected molecular mass of 33 ku. The expression levels of these two recombinant proteins accounted for more than 20% of the total cellular proteins (Figure 2). The yields were approximately 32 and 50 mg/L for A2-BSP and β2m, respectively. Western blotting showed that the recombinant β2m could react with antibodies against human native β2m (data not shown). HLA-A2 heavy chain expression was not analyzed by Western blotting, because no suitable antibody was available. Both A2-BSP and β2m were largely expressed in the insoluble fraction (inclusion bodies), which facilitated purification through simple washing and centrifugation (Figure 3, lanes 2 and 3).
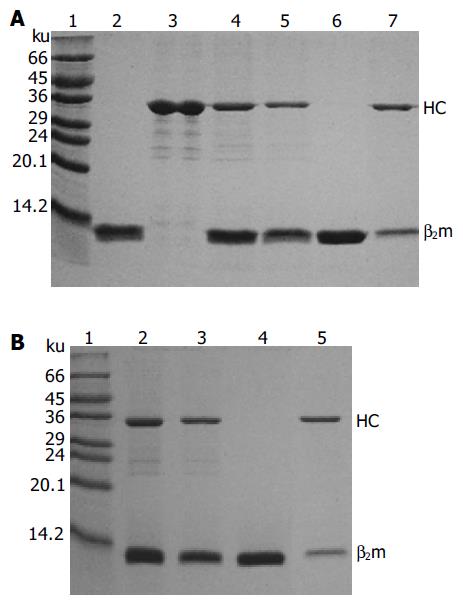
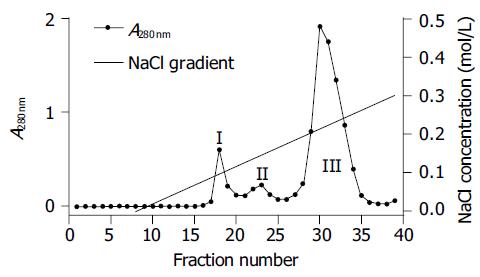
Monomeric HLA-A2 was refolded by dilution of A2-BSP and β2m with refolding buffer in the presence of HLA-A*0201-restricted antigenic peptides. Two peptides were used to reconstitute the HLA-A2 complex. One was the immunodominant NLVPMVATV (NLV) peptide, which was derived from pp65 (pp65495-503) of HCMV[11] and used to reconstitute the HLA-A2-NLV (hereafter referred to as A2-NLV) complex. The other was the predominant CTL epitope GILGFVFTL (GIL) peptide derived from the matrix protein (Mp58-66) of influenza A virus[15], which was used to reconstitute the HLA-A2-GIL (referred to as A2-GIL) complex. The yield of refolding was about 10-15%. The refolded A2-NLV and A2-GIL complexes appeared as two bands (A2-BSP and β2m) on SDS-PAGE (Figure 3). After biotinylation, each complex was purified by single Q-Sepharose column chromatography. Typically, three peaks eluted from the column, as shown by monitoring of absorbance at 280 nm (Figure 4). Soluble β2m eluted as the first peak. The second peak comprised the desired soluble HLA-A2 complex (A2-NLV or A2-GIL) consisting of a heavy chain and β2m (Figure 3), which was of 95% purity as analyzed with the PhotoCapt Ver11.01 software (Vilber Lourmat, France). The fractions in this peak were pooled and concentrated by ultrafiltration. The third peak contained non-protein materials with a much higher absorbance at 260 nm than at 280 nm. Refolding without peptide (negative control) yielded no stable HLA-A2 complex.
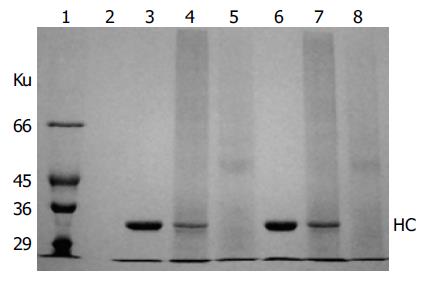
HLA-A2 tetramers were formed by mixing the biotinylated A2-NLV or A2-GIL monomers with streptavidin-PE at a 4:1 molar ratio. SDS-PAGE analysis of the HLA-A2 tetramers could be used to estimate the extent of tetramer formation, because the complex formed between streptavidin and biotin was stable in the absence of boiling treatment and DTT[10]. As shown in Figure 5, the single main band observed in the lanes of monomeric A2-NLV or A2-GIL corresponded to the 33 ku heavy chain, as β2m quickly migrated to the bottom of the gel. Software analysis (PhotoCapt Ver11.01) showed that the amount of heavy chain in the A2-NLV tetramer lane was about 10-20% of that in the lane containing monomeric HLA-A2, indicating that about 80% of the A2-NLV monomer formed multimers with streptavidin-PE. The A2-GIL tetramer showed a similar extent of multimer formation.
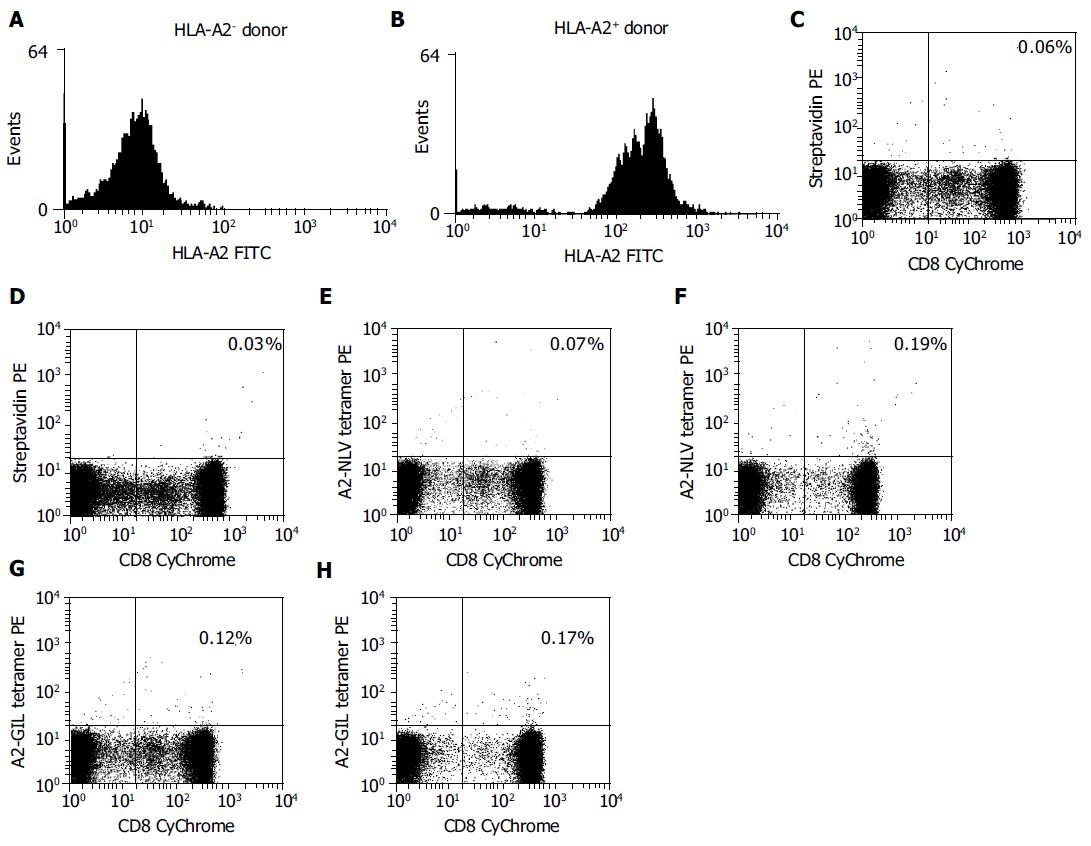
Finally, the A2-NLV and A2-GIL tetramers were tested by flow cytometry for their abilities to identify HCMV- or influenza-specific CD8+ T cells in freshly isolated PBMCs from HLA-A2 positive and negative donors. PBMCs were analyzed by three-color flow cytometry after A2-NLV and A2-GIL tetramer staining. Background staining was performed with streptavidin-PE instead of the tetramers.
Subpopulations of CD8+ T cells specific for either pp65495-503 or Mp58-66 epitopes were found in HLA-A2 positive donors by A2-NLV or A2-GIL tetramer staining. Of the total T cells, 0.19% were stained by the A2-NLV tetramer, while 0.17% bound to the A2-GIL tetramer (Figure 6). However, only 0.07% and 0.12% of T cells from the HLA-A2 negative donor were nonspecifically stained by the A2-NLV or A2-GIL tetramers, respectively, which was comparable to the control staining with streptavidin-PE (0.06%). This result suggests that these tetramers are quite specific for MHC-restricted T cells.
As the current method of limiting dilution assay (LDA) tends to underestimate CTL frequency[2,3], other methods have been sought for the quantification of antigen-specific CD8+ T cells in immune responses. T cell antigen receptors (TCR) expressed on CD8+ T cells can recognize peptide-MHC complexes, the specific ligands on the antigen-presenting cells (APCs). Therefore, direct staining of CD8+ T cells with their soluble cognate ligands should be an ideal approach. However, the strategy initially failed because soluble monomeric peptide-MHC complexes have low affinities for the T-cell receptor (TCR)[1-3]. This obstacle has been overcome by Altman et al.[3]. Tetramer reagents have been shown to bind specifically to cognate T cells in numerous systems, allowing fast and direct quantification of antigen-specific T cells by flow cytometry[3,4,8,9,18-20]. Thus, tetramer-based assays have become a powerful new technology for detecting antigen-specific T cells[4,8,21]. However, the general application of this strategy is limited by the complex, labor intensive protocols required for preparing specific peptide-MHC tetramers.
It is difficult to prepare a large amount of MHC class I heavy chain, because the translation efficiency in E. coli is very low. However, optimization of codons in the TIR has been shown to increase translation efficiency[14]. Sato et al.[12], found that the expression of wild type HLA-A*2402 heavy chain in E. coli is undetectable, whereas a synonymous mutant in which the TIR mammalian usage codons are replaced by those of E. coli, can be expressed at high levels. Lakey et al.[22], showed that the production of recombinant mycobacterial proteins increases 54-fold following selective replacement of low-usage E. coli codons in mycobacterial proteins with high-usage E. coli codons. In addition, the high G/C content in the TIR can block the expression of human proteins in E. coli[14]. The previous paper did not include a detailed description of the expression vector or E. coli system, instead of referring readers to the system used by Garboczi[17], in which no codon or G/C content changes were described. In the present study, recombinant proteins were expressed largely in the form of inclusion bodies, which greatly facilitated isolation and purification of the refolded peptide-MHC complexes, thus simplifying the tetramer preparation procedure. In addition to the difficulty in obtaining a large amount of MHC heavy chain, the practical application of tetramers is also limited by the complexity of the necessary preparation procedures. In this study, we reconstituted the monomeric HLA-A2 complexes based on the dilution strategy described by Garboczi[17]. After biotinylation, the HLA-A2 monomer could be highly purified by single step ion-exchange chromatography. It should be noted that the purification in the present study was carried out in a low pressure chromatography system requiring no HPLC facilities, whereas the previous protocol included three purification steps with high performance column chromatography[3].
The results of this study indicate that the generated reagents can be used for the detection of antigen-specific T cells. Our novel procedure for generating tetramers should be applicable for preparing other HLA-A2 tetramers or for generating tetramers of other human MHC class I alleles.
Tetramer technology is primarily used to determine the frequency of antigen-specific T cells[3-5], and has been extensively used in evaluating the dynamics of cytomegalovirus (CMV)-specific CTL responses in humans[5-7,9,11]. It is generally believed that adults over 40 years in developing countries are almost 100% CMV positive[9], and that the pp65495-503 NLV peptide (NLVPMVATV) is the predominant CMV epitope presented by HLA-A2 molecules[11,23]. Quantification of specific T cells in the peripheral blood of healthy CMV carriers using an A2-NLV tetramer demonstrated that the percentage of NLV-specific CTLs in CMV carriers is about 0.02-6.19% within the CD8+ T cell population[24]. In our study, tetramer staining showed that there were 0.19% NLV-specific CTLs among total T cells from the peripheral blood of a randomly selected HLA-A2-positive volunteer over 40 years, confirming again that NLV dominates the HLA-A2-restricted cellular immune response against CMV[23]. There is substantial evidence that quantification of NLV-specific CTLs by tetramers is of clinical significance[5-7,9,11,24,25]. Despite antiviral therapy, CMV remains an important cause of morbidity and mortality after allogeneic stem cell transplantation (SCT)[26]. Through direct quantification of CMV-specific CD8+ T cells using A2-NLV tetramers, several studies demonstrated that recovery of CMV-specific CD8+ T cells after SCT is critical for protection against CMV disease[6,7,25]. Accordingly, enumeration of HLA-restricted, CMV-specific CD8+ T cells by tetramer technology in grafts, and monitoring of these cells after SCT may constitute a rapid and sensitive tool for identifying SCT recipients at risk for developing CMV disease. Hence, tetramer technology is of value both in the development of novel transplant protocols and in clinical management of individual cases[5-7,9,25].
It is also possible to combine tetramer staining with other flow cytometry-based assays to gain more phenotypic or even functional information on antigen-specific CTLs[3,8,10,12]. For example, tetramer staining can be combined with intracellular detection of cytokines (e.g. interferon-γ), chemokines (e.g. macrophage inflammatory protein 1α), and cytotoxins (e.g. perforin/granzymes) following in vitro antigen/mitogen stimulation to assess the functional status of tetramer-positive T cells[27,28]. Moreover, tetramers can be used in the isolation of antigen-specific cells for characterization and even expansion of immunotherapeutic use[11,25,29]. Conjugation of a specific tetramer with antibodies against a tumor-associated antigen has been used to redirect CMV-specific CTLs to kill tumor cells by binding to the surface of tumor cells and sensitizing them to lysis by tetramer-specific CTLs, suggesting the potential therapeutic application of tetramers[30]. Thus, tetramer technology has opened new routes for further study of T-cell responses and tumor immunotherapy.
In summary, we have established a simplified and efficient procedure for generating peptide-MHC tetramers, a highly specific and very useful reagent with a number of important applications in cellular immune response studies. The present method may provide an alternative method for the preparation of MHC class I tetramers, and promote the use of this exciting technology.
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annu Rev Immunol. 2000;18:275-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 534] [Cited by in RCA: 534] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 2. | Doherty PC, Christensen JP. Accessing complexity: the dynamics of virus-specific T cell responses. Annu Rev Immunol. 2000;18:561-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 221] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2847] [Cited by in RCA: 2867] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 4. | Meidenbauer N, Hoffmann TK, Donnenberg AD. Direct visualization of antigen-specific T cells using peptide-MHC-class I tetrameric complexes. Methods. 2003;31:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Gratama JW, Cornelissen JJ. Diagnostic potential of tetramer-based monitoring of cytomegalovirus-specific CD8+ T lymphocytes in allogeneic stem cell transplantation. Clin Immunol. 2003;106:29-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Gratama JW, van Esser JW, Lamers CH, Tournay C, Löwenberg B, Bolhuis RL, Cornelissen JJ. Tetramer-based quantification of cytomegalovirus (CMV)-specific CD8+ T lymphocytes in T-cell-depleted stem cell grafts and after transplantation may identify patients at risk for progressive CMV infection. Blood. 2001;98:1358-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 225] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Cwynarski K, Ainsworth J, Cobbold M, Wagner S, Mahendra P, Apperley J, Goldman J, Craddock C, Moss PA. Direct visualization of cytomegalovirus-specific T-cell reconstitution after allogeneic stem cell transplantation. Blood. 2001;97:1232-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 230] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 8. | Xu XN, Screaton GR. MHC/peptide tetramer-based studies of T cell function. J Immunol Methods. 2002;268:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Lacey SF, Diamond DJ, Zaia JA. Assessment of cellular immunity to human cytomegalovirus in recipients of allogeneic stem cell transplants. Biol Blood Marrow Transplant. 2004;10:433-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 10. | Bousso P. Generation of MHC-peptide tetramers: a new opportunity for dissecting T-cell immune responses. Microbes Infect. 2000;2:425-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | Bodinier M, Peyrat MA, Tournay C, Davodeau F, Romagne F, Bonneville M, Lang F. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat Med. 2000;6:707-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 125] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Sato Y, Sahara H, Tsukahara T, Kondo M, Hirohashi Y, Nabeta Y, Kawaguchi S, Ikeda H, Torigoe T, Ichimiya S. Improved generation of HLA class I/peptide tetramers. J Immunol Methods. 2002;271:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54:198-210. [PubMed] |
| 14. | Song ZL, Pan QH, Wang J, Yang Y, Zhu DM, Chen XJ, Liu CL, Hong A. Cloning and high expression of hbFGF with a new strategy. Yi Chuan Xue Bao. 2002;29:84-89. [PubMed] |
| 15. | Brosterhus H, Brings S, Leyendeckers H, Manz RA, Miltenyi S, Radbruch A, Assenmacher M, Schmitz J. Enrichment and detection of live antigen-specific CD4(+) and CD8(+) T cells based on cytokine secretion. Eur J Immunol. 1999;29:4053-4059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | He XH, Xu LH, Liu Y, Zeng YY. Cloning of human beta-microglobulin gene and its high expression in Escherichia coli. ShengWu GongCheng XueBao. 2004;20:99-103. [PubMed] |
| 17. | Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA. 1992;89:3429-3433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 585] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 18. | Trudeau JD, Kelly-Smith C, Verchere CB, Elliott JF, Dutz JP, Finegood DT, Santamaria P, Tan R. Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J Clin Invest. 2003;111:217-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 193] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 19. | Skinner PJ, Haase AT. In situ tetramer staining. J Immunol Methods. 2002;268:29-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Ferrari G, Neal W, Ottinger J, Jones AM, Edwards BH, Goepfert P, Betts MR, Koup RA, Buchbinder S, McElrath MJ. Absence of immunodominant anti-Gag p17 (SL9) responses among Gag CTL-positive, HIV-uninfected vaccine recipients expressing the HLA-A*0201 allele. J Immunol. 2004;173:2126-2133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Gratama JW, Kern F. Flow cytometric enumeration of antigen-specific T lymphocytes. Cytometry A. 2004;58:79-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Lakey DL, Voladri RK, Edwards KM, Hager C, Samten B, Wallis RS, Barnes PF, Kernodle DS. Enhanced production of recombinant Mycobacterium tuberculosis antigens in Escherichia coli by replacement of low-usage codons. Infect Immun. 2000;68:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, Sissons JG. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569-7579. [PubMed] |
| 24. | Hassan-Walker AF, Vargas Cuero AL, Mattes FM, Klenerman P, Lechner F, Burroughs AK, Griffiths PD, Phillips RE, Emery VC. CD8+ cytotoxic lymphocyte responses against cytomegalovirus after liver transplantation: correlation with time from transplant to receipt of tacrolimus. J Infect Dis. 2001;183:835-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 25. | Einsele H, Roosnek E, Rufer N, Sinzger C, Riegler S, Löffler J, Grigoleit U, Moris A, Rammensee HG, Kanz L. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood. 2002;99:3916-3922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 547] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 26. | Broers AE, van Der Holt R, van Esser JW, Gratama JW, Henzen-Logmans S, Kuenen-Boumeester V, Löwenberg B, Cornelissen JJ. Increased transplant-related morbidity and mortality in CMV-seropositive patients despite highly effective prevention of CMV disease after allogeneic T-cell-depleted stem cell transplantation. Blood. 2000;95:2240-2245. [PubMed] |
| 27. | Appay V, Nixon DF, Donahoe SM, Gillespie GM, Dong T, King A, Ogg GS, Spiegel HM, Conlon C, Spina CA. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J Exp Med. 2000;192:63-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 716] [Cited by in RCA: 713] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 28. | Lichterfeld M, Yu XG, Waring MT, Mui SK, Johnston MN, Cohen D, Addo MM, Zaunders J, Alter G, Pae E. HIV-1-specific cytotoxicity is preferentially mediated by a subset of CD8(+) T cells producing both interferon-gamma and tumor necrosis factor-alpha. Blood. 2004;104:487-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Keenan RD, Ainsworth J, Khan N, Bruton R, Cobbold M, Assenmacher M, Milligan DW, Moss PA. Purification of cytomegalovirus-specific CD8 T cells from peripheral blood using HLA-peptide tetramers. Br J Haematol. 2001;115:428-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Robert B, Guillaume P, Luescher I, Romero P, Mach JP. Antibody-conjugated MHC class I tetramers can target tumor cells for specific lysis by T lymphocytes. Eur J Immunol. 2000;30:3165-3170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |