INTRODUCTION
Helicobacter pylori (H pylori) is highly associated with active and chronic gastritis as well as peptic ulcer diseases. H pylori is also considered to be a class I carcinogen because of its role in the development of gastric carcinoma and gastric mucosa-associated lymphoid tissue lymphoma. In addition to gastric Helicobacters, Helicobacter species have been isolated from the intestinal tract, liver and bile ducts of animals, and they have also been found to play a pathological role in enterohepatic diseases in animals[1]. Recently, several separate research groups have detected such Helicobacter DNA as H pylori, H bilis, Flexispira rappini and H pullorum in bile, gall bladder or liver tissue obtained from patients with chronic cholecystitis, primary sclerosing cholangitis (PSC), primary biliary cirrhosis (PBC), or primary liver carcinoma[2-12]. Our previous study showed that Helicobacter species DNA could be found in liver tissue from 52.6% to 60% patients with primary liver carcinoma by polymerase chain reaction (PCR) using 16S rRNA primers, while none was found in control group. PCR amplicons were identified by Southern blot and sequenced, and the homology similarity was 99-100% with 16S rRNA of H pylori[13-15]. These reports suggested that Helicobacter organisms, including H pylori, may play a pathogenic role in the development of hepatobiliary diseases in humans. Whether Helicobacter spp. is an innocent bystander, cofactor or culprit, it is still under review[16].
Proteome is a new word that was proposed by Wilkins[17] to define all the different proteins occurring in an organism in space and time. Proteomics is an emerging area of research of the post-genomic era which is based on three technological platforms, high-resolution two-dimensional gel electrophoresis (2-DE), highly sensitive biological mass spectrometry (MS), and the rapidly growing protein and DNA databases. During the past few years, proteomics has been extensively applied to several fields of medicine to better understand normal physiology, to define the pathophysiology of diseases, and to identify novel biomarkers and new therapeutic targets[18]. In order to further explore the pathological effect of H pylori on human hepatic cells, we have employed high-throughput and high-resolution 2-DE analysis of human HepG2 cell extracts to examine protein expression patterns between untreated and H pylori-treated HepG2 cells, and certain differential expression proteins were identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) and database analysis. The results presented here will no doubt provide clues to study further the molecular mechanisms of pathological effect of H pylori on hepatic cells.
MATERIALS AND METHODS
Chemicals and materials
Immobilized pH gradient (IPG) strips as well as ImageMaster 2D Elite 3.01 software were purchased from Amersham Pharmacia Biotechnology (Uppsala, Sweden). Acrylamide and other reagents for the polyacrylamide gel preparation were from Amresco (OH, USA); urea, CHAPS, thiourea, DTT, iodoacetamide, proteomics grade trypsin, CCA and antibiotics were from Sigma (St. Louis, MO, USA); medium and fetal bovine serum (FBS) for cell culture were from Gibco-BRL (Grand Island, NY, USA).
Methods
Cell line and cell culture The human hepatic cell line HepG2 was purchased from Cell Bank of Shanghai Academy of Science. The cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 100 mL/L FBS, 100 U/mL penicillin G, and 100 μg/mL streptomycin sulfate in an atmosphere of 50 mL/L CO2 at 37 °C. The cells were maintained in the logarithmic growth phase for stimulation experiment.
H pylori strains and culture
H pylori reference strain ATCC49503 (cagA+, vacA s1a) was used in the experiment. The bacteria were plated onto Columbia agar containing 70 mL/L sheep blood, 10 μg/mL vancomycin, 2 μg/mL amphotericin B, and 20 μg/mL polymyxin B sulfate and incubated at 37 °C under microaerobic conditions[19]. After 3 d, the bacteria were harvested into 0.01 mol/L phosphate-buffer saline (PBS) and pelleted by centrifugation at 4000 r/min for 10 min, and resuspended in antibiotic-free DMEM with 100 mL/L FBS to a concentration of 1×108 CFU/mL.
Co-culture of H pylori and HepG2 cells and protein extraction
For all experiments, H pylori (1×108 CFU) were added to HepG2 (1×106) at a bacteria:cells concentration of 100:1. Control cells were incubated with DMEM/100 mL/L FBS alone, and co-incubation was performed up to 6 h in triplicate. At the end of incubation, the majority of adherent bacteria were removed by several washes in ice-cold PBS. The cells were collected by trypsinization followed by centrifugation. Finally, the cells were lysed in 200 μL lysis buffer (8 mol/L urea, 2 mol/L thiourea, 40 g/L CHAPS, 40 mmol/L Tris, 65 mmol/L DTT, 5 mmol/L PMSF). The lysates were vortexed followed by incubation at room temperature for 30 min and then centrifuged at 15000 g for 30 min at 4 °C. The supernatant was transferred to a fresh tube and the concentration of the total protein was determined by the Bradford method. Aliquots were stored at -80 °C until use.
2-DE
2-DE was performed as described[20]. The first dimensional isoelectric focusing (IEF) was performed on precast 18-cm immobilized pH 3-10 gradient (IPG) strips at 20 °C using a IPGphor IEF Cell (Amersham Biosciences). Typically, 300 μg of total protein were mixed with a rehydration buffer containing 8 mol/L urea, 20 g/L CHAPS, 3 g/L DTT, 5 mL/L IPG buffer (pH 3-10) and a trace of bromophenol blue, to a total volume of 350 µL. The mixtures were loaded onto the IPG strip and focusing was carried out for 41 920 Vh. Following IEF, the gel strip was first equilibrated for 15 min in the equilibration buffer containing 50 mmol/L Tris-HCl (pH 8.8), 6 mol/L urea, 300 mL/L glycerol, 20 g/L SDS, 2 g/L DTT, and a trace of bromophenol blue. Then the gel strip was equilibrated for another in the same equilibration buffer, except that DTT was replaced with 30 g/L iodoacetamide. Second-dimensional SDS-PAGE was performed in 12.5% acrylamide gels using the ProTEAN II system (Bio-Rad) as described by manufacturer and Gorg[20]. After electrophoresis, the protein spots were visualized by silver nitrate.
Image analysis
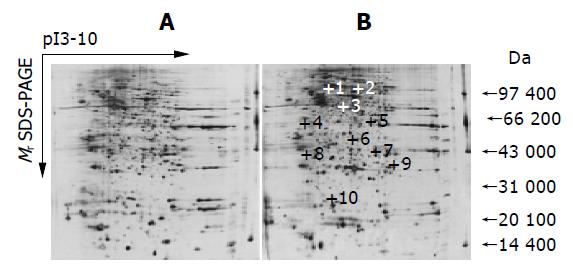
The stained 2-DE gels were scanned with LabScan software on Imagescanner (Amersham Biosciences). ImageMaster 2D Elite 3.01 analysis software was used for spot-intensity calibration, spot detection, background abstraction, matching, and 1-D calibration. The reproducibility of spot position was calculated with Corbett’s method[21]. Statistical analysis was carried out with SPSS for Windows 10.0 and Excel. To account for experimental variations, three gels were prepared for each group. The gel spot pattern of each gel was summarized in a standard after spot matching. Thus, we obtained one standard gel for each group. These standards were then matched to yield information about differences in abundance related to H pylori treatment (up- or down regulation of spots).
MALDI-TOF-MS
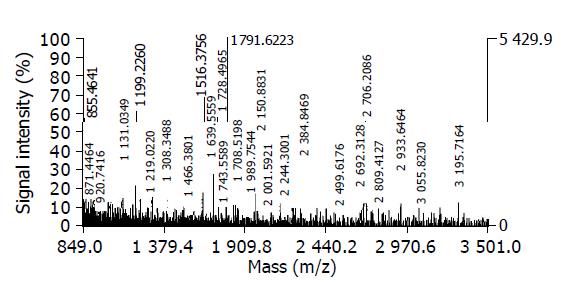
Protein spots were excised from preparative gels using biopsy punches and transferred to a 1.5-mL siliconized Eppendorf tube. Proteins were in gel digested as previously described[22]. The gel spots were destained in the destaining solution that consisted of 100 mmol/L Na2S2O3 and 30 mmol/L K3Fe(CN)6 (V/V, 1:1). The protein-containing gel spots were reduced in the reduction buffer (100 mmol/L NH4HCO3 and 10 mmol/L DTT) for 1 h at 57 °C and then alkylated in the alkylation buffer (100 mmol/L NH4HCO3 and 55 mmol/L iodoacetamide) in the dark for 30 min at room temperature. The gel pieces were dried in a vacuum centrifuge. The dried gel pieces were incubated in the digestion solution that consisted of 40 mmol/L NH4HCO3 and 20 μg/mL proteomics grade trypsin for 24 h at 37 °C. The peptide digest (1 μL) was mixed with 1 μL CCA matrix solution and 1 μL of this mixture was applied to the stainless steel plate and air dried. The samples were analyzed with Voyager DE STR MALDI-TOF Mass Spectrometer (Applied Biosystems, Cambridge, MA). The parameters were set up as follows: positive ion-reflector mode, accelerating voltage 20000 V, grid voltage 68%, mirror voltage ratio 1.12, N2 laser wavelength 337 nm, pulse width 3 ns, the number of laser shots 50, acquisition mass range 900-3500 Da, delay 180 ns, and vacuum degree 4×10-7 Torr. A list of the corrected mass peaks was the peptide mass fingerprinting (PMF).
Database analysis
Proteins were identified with PMF data by searching software PeptIdent (http://www.expasypku.edu.cn). The searching parameters were set up as follows: acquisition mass ranges 900-3500 Da, the mass tolerance was ±0.5 Da; the number of missed cleavage sites was allowed up to 1; the cysteine residue was modified as carbamidomethyl-cys; the minimum number of matched peptide was four; species was selected as Homo sapiens (humans); the peptide ion was [M+H]+; the isotope masses were used; and the searching range was within the experimental pI value±0.5 pH unit and experimental Mr±20%.
DISCUSSION
The detection that Helicobacter spp. could infect biliary tract and liver of humans has been reported by several studies in different settings. Fox et al[6], have shown the presence of Helicobacter spp. in the bile of Chileans with chronic cholecystitis. Avenaud and coworkers[3] have demonstrated by PCR the presence of genomic sequences of Helicobacter spp. in the liver of eight patients with hepatocellular carcinoma (HCC) without primary diagnosis of cirrhosis, and a further analysis by sequencing revealed that these species were H pylori and H felis. Similarly, Ponzetto[12] has reported the presence of cagA gene sequences obtained from the liver tissue of cirrhotic patients with HCC. Furthermore, Nilsson et al[2], have identified H pylori and Helicobacter spp. in human liver samples from patients suffering from PSC and PBC and recently in liver samples of patients with cholangiocarcinomas or HCC[5]. Of great interest is the fact that the gene sequence obtained from positive PCR of Helicobacter spp.-specific 16S rRNA was usually most analogous to H pylori. Helicobacter spp. has been successfully cultured from the liver of a patient with Wilson’s disease by Queiroz and Santos[23]; the isolate is closely related to H pylori by biochemical and 16S rRNA analysis. Lately, Wang et al[24], followed-up C57BL/6 and Balb/cA mice infected by three different H pylori isolates. They found that one C57BL/6 mouse infected with H pylori strain 119p developed HCC after 23 mo. Taking the results together, we may conclude that, as in animals, Helicobacter species especially H pylori may colonize the liver of humans, which could have some sort of importance in the pathogenesis and in the development of some liver diseases.
The comparative proteomic study was performed between the untreated and H pylori-treated HepG2. Ten differential protein spots were selected to perform in-gel trypsin digestion and MALDI-TOF-MS-based PMF analysis. Seven of these proteins were successfully identified by MALDI-TOF-MS. Some of the identified proteins are the oncogene-encoded products and others are the regulatory proteins of transcription and signal transduction.
The integrins are a family of cell surface glycoproteins which mediated interactions of cells with the extracellular matrix and cells with cells. These integrin-mediated interactions influence many aspects of cell behavior including cell survival, cell adhesion, cell migration, as well as cell proliferation and differentiation. All integrins are heterodimers composed of two subunits, α and β. Integrin β-1 has been found to play an essential role in invasion and metastasis of HCC cells[25]. Liu et al, found that integrin β-1 gene was upregulated in HCC using cDNA microarray technology, which was consistent with RT-PCR and Northern blot analysis of integrin β-1 gene. Human hepatoma cell lines HepG2, Huh7, and HLE stably transfected with full-length integrin β-1 had overexpression of integrin β-1 and were protected from apoptosis induced by chemotherapeutic reagents via a mitogen-activated protein kinase (MAPK)-dependent pathway[26].
Protein kinase C (PKC) is a family of at least 11 structurally related phospholipid-dependent serine/threonine kinases and it plays crucial roles in transducing signals that regulate diverse cellular functions, including proliferation, differentiation, and apoptosis. The PKC family is divided into three subgroups based on differences in their structures and co-factor requirements. PKC-α is a member of the group A or classical PKCs (cPKCs). Review of the literatures has shown that the PKC signal transduction pathway plays an important role in the development of many diseases and, in particular, cancer[27]. PKC-α is an important tumor-promoting factor; as potential target for anticancer therapy, PKC-α is currently being studied[28].
Homeobox genes play fundamental roles in development. They can be subdivided into several subfamilies, one of which is the LIM-homeobox subfamily. The characteristic features of LIM-homeodomain proteins are double zinc finger motifs, called LIM domains, that are located N-terminally of the homeodomain. These proteins mediate protein-protein interactions and are of fundamental importance for cell differentiation, cytoskeletal remodeling and transcriptional regulation[29].
Eukaryotic translation initiation factor 2 (eIF-2) is a multimeric protein consisting of three dissimilar subunits termed α, β, and γ, which functions as a central switch in the initiation of protein synthesis. In its GTP-bound state it delivers the methionyl initiator tRNA (Met-tRNA(i)) to the small ribosomal subunit and releases it upon GTP hydrolysis following the recognition of the initiation codon[30]. PINCH protein is an effector of integrin and growth factor signaling, coupling surface receptors to downstream signaling molecules involved in the regulation of cell survival, cell proliferation and cell differentiation[31].
MAPK pathway is an important signal transduction pathway that transduces extracellular signals into intracellular responses and has been implicated in a wide array of physiological processes such as proliferation, differentiation, development, apoptosis, stress and inflammatory responses. The MAPK kinases (MAPKKs) are up-stream dual-specificity protein kinases which phosphorylate and activate MAPK on threonine and tyrosine residues[32].
Ras-related protein Rab-37 belongs to the small GTPase superfamily, Rab family. GTPases regulate a myriad of cellular functions including signal transduction, cytoskeletal organization and membrane trafficking. Rab family members are also widely involved in human tumorigenesis by overexpression[33,34].
The upregulation of these proteins suggests that the capacity of the tumor invasion is stronger. However, the exact mechanisms through which H pylori lead to the change of these proteins await further study.
Advances in 2D electrophoresis, MS, and bioinformatics now make possible the large-scale examination of protein expression patterns. These improved methods are used to identify proteins found to be upregulated in expression level in H pylori-treated HepG2 cells. The above findings could be valuable for further elucidating the etiological role of Helicobacter spp. infection in a variety of liver disorders.