Published online Jun 14, 2005. doi: 10.3748/wjg.v11.i22.3363
Revised: May 28, 2004
Accepted: July 11, 2004
Published online: June 14, 2005
AIM: To construct and highly express an epitope of hepatitis C virus (HCV) in a foreign epitope presenting vector based on an insect virus, and to study the antigenicity of the epitope.
METHODS: The HCV epitope sequence (amino acid residues 315 to 328: EGHRMAWDMMMNWS) of the E1 region was constructed at different positions of a foreign epitope presenting vector based on an insect virus, flock house virus (FHV) capsid protein encoding gene as a vector, and expressed in E. coli cells. Western blotting and ELISA were used to detect the immunoreactivity of these recombinant proteins.
RESULTS: The gene encoding of the concerned B-cell epitope of HCV E1 envelope protein was expressed on FHV capsid carrier protein at positions I1 (aa 106), I2 (aa 153) and I3 (aa 305), respectively, on the surface of FHV capsid protein. The recombinant proteins in this system could be highly expressed in more than 40% of total cell protein of E. coli BL21. All the expressed recombinant proteins were in inclusion body form, and showed obvious immunoreactivity by Western blotting. Further purified recombinant proteins were detected by indirect ELISA as coating antigen respectively. All recombinant proteins could still show immunoreactivity.
CONCLUSION: The epitope of HCV E1 envelope protein can be highly expressed in FHV carrier system as a chimeric protein with high immunoreactivity. This system has multiple entry sites conferring many possible conformations closer to the native one for a given sequence.
- Citation: Peng M, Dai CB, Chen YD. Expression and immunoreactivity of an epitope of HCV in a foreign epitope presenting system. World J Gastroenterol 2005; 11(22): 3363-3367
- URL: https://www.wjgnet.com/1007-9327/full/v11/i22/3363.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i22.3363
Hepatitis C virus is the major etiological agent for non-A, non-B hepatitis[1-3]. There are 1.7 hundred million infected patients all over the world[4], and chronic infection is established in about 50-90% of patients[5]. HCV-infected patients develop a heterogeneous immune response. The initial worldwide screening of blood for antibodies against HCV was carried out with a polyprotein (c100-3) from the NS3-NS4 region[6], but the specificity and sensitivity of this region are not sufficient[7,8]. Recent studies have identified several epitopes in the envelope protein[9-12].
The characterization of the sequences recognized by antibodies and their reactivity has been performed mainly with the use of synthetic peptides[13,14]. However, recent studies have provided evidence that there are severe limitations in the use of synthetic peptides bound to the solid phase for the detection of antibodies against conformational epitopes[15,16]. A new epitope presenting system, based on the capsid protein of the flock house virus (FHV), has been successfully used in the presentation of several conformational epitopes of human immunodeficiency virus type 1[17]. Therefore, we inserted the FHV capsid protein in three portions of an epitope of HCV envelope protein, and studied the immunogenicity of these chimeric proteins.
HCV patient serum samples were kindly provided by Kunming Infectious Diseases Hospital. Antisera from HCV-Eb chimeric protein (spanning hole HCV E1 envelope protein without FHV capsid carrier protein) immunized guinea pigs were prepared in our laboratory.
The studied epitope was the E1 region of HCV comprised of 14 amino acids, EGHRMAWDMMMNWS, from the residues 315-328. These residues had the highest conservation in E1 region. Two primers coding this epitope were synthesized by Sangon Company: primers Ps (5’-tgagggtc-accgtatggcttgggacatgatgatgaactggtctcc-3’) and Pas (3’-cccagtggcataccgaaccctgtcctactacttgaccagaggagt-5’).
The annealing reaction mixture containing 4 μL primer Ps (≈100 ng), 4 μL primer Pas (≈100 ng), 5 μL buffer (10×) and 12 μL H2O was incubated at 65 °C for 5 min, then cooled down to 37 °C for 45 min in 37 °C water bath. The annealed mixture was added with 2 μL ATP (25 mol/L), 2 μL kinase (10 U/μL) and 3 μL buffer (10×), and incubated in a 37 °C water bath for 1 h.
FHV capsid gene RNA2 was genetically modified, having a restriction endoenzyme site Bsu36I at different positions of the molecule. The FHV-RNA2 was constructed onto the plasmid pET-3, and the product was designated as pET-RNA2.
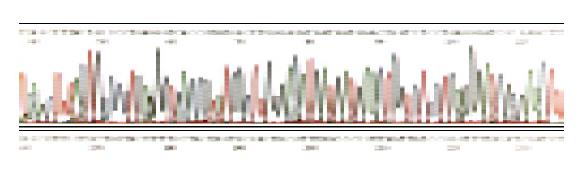
After digestion with Bsu 36I (purchased from Biolabs), pET-RNA2 as a vector was ligated with kinased HCV epitope primers (double stranded), and transformed in E.coli DH5α competent cells. The positive recombinant plasmids were identified by digestion with proper restriction endoenzymes respectively, and finally sequenced by the dideoxy chain determination method with T7 DNA polymerase (T7 sequencingTM, Pharmacia Biotech Inc., USA). Then correct plasmids was identified, designated as pET-RNA2-E1, and used for recombinant epitope (chimeric proteins) expression.
Competent BL21 (DE3) was transformed with the recombinant plasmid pET-RNA2-E1, and incubated in LB medium. After transformation and incubation, 3 mL fresh culture was transferred into 250 mL fresh TB-P medium (phosphate-rich medium, containing 200 μg/mL ampicillin), and incubated overnight. The cells were then gathered by low-speed centrifugation and resuspended in 50 mL of sonication buffer. After sonication lysis and centrifugation, the recombinant epitope/chimeric protein was obtained in inclusion body form. The expressed proteins were detected in 120 g/L SDS-PAGE gels.
Total cell lysates were run on SDS-PAGE gels and transferred electrophoretically to nitrocellulose membrane for 2 h at the voltage of 100 V. The membrane was then incubated in blocking solution (50 g/L nonfat milk in Tris-buffered saline, TBS) for 1 h at room temperature at 80 r/min followed by incubation at room temperature for 2 h in the HCV positive sera prediluted to 1:100 with blocking solution. The membrane was washed thrice with TBS/T (1 g/L Tween-20 in TBS) for 10 min, and horseradish peroxidase-labeled goat anti-human IgG antibodies (purchased from Sigma) diluted in TBS/T (1:2000) were exposed to the membrane at room temperature for 1 h. The membrane was visualized with a substrate solution of DAB (purchased from Sigma) and NiCl2 after washing thrice for 10 min with TBS/T.
ELISA for recombinant protein of HCV E1 epitope and peptide of the E1 epitope was done in 96-well, flat-bottomed vinyl assay plates. Microplates were coated with purified recombinant protein or synthetic HCV E1 peptide in 0.05 mol/L sodium carbonate buffer (pH 9.6) for 2 h at 37 °C and overnight at 4 °C. The recombinant protein was diluted to 0.5 μg/mL for ELISA, and the peptide was diluted to 5 μg/mL. Plates were washed 4 times with PBS containing 0.5 g/L Tween 20 and blocked with blocking buffer (0.5 g/L Tween 20, 2.5 g/L bovine serum albumin and 0.5 g/L NaN3 in PBS) for 2 h at 37 °C. Antisera against HCV-Eb (1:1000) were applied for 30 min at 37 °C. A peroxidase-conjugated goat anti-guinea pig IgG used as secondary antibody was incubated for 30 min at 37 °C. Wells were washed four times with PBS/T between each step and visualized with o-phenyl-diamine-2HCL (50 mg/L in PBS, pH 5.0). The reaction was stopped with 50 μL of 2 mol/L H2SO4. Absorption was measured at A495.
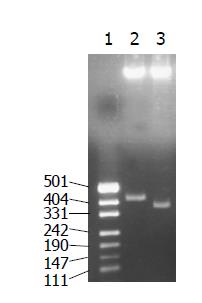
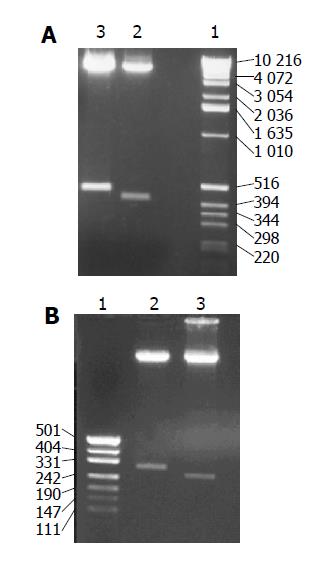
The recombinant plasmids were identified by digestion with proper restriction endoenzymes, respectively. A 410 bp (including 45 bp E1 epitope gene) fragment was obtained when pET-RNA-I1-E1 was digested with SpeI/MluI, a 365 bp fragment was obtained when pET-RNA-I1-wt digested with the same endoenzymes; A 472 bp (including 45 bp E1 epitope gene) fragment was obtained when pET-RNA-I2-E1 digested with SpeI/NcoI, a 427 bp fragment was obtained when pET-RNA-I2-wt digested with SpeI/NcoI; A 302 bp (including 45 bp E1 epitope gene) fragment was obtained when pET-RNA-I3-E1 digested with NsiI/NcoI, a 257 bp fragment was obtained when pET-RNA-I3-wt digested with the same endoenzymes; agarose gel electrophoresis showed that the HCV epitope gene was cloned into three positions of the pET-RNA2 vector with correct size, (Figures 1 and 3). The nucleotide sequence of inserted fragments were identified by sequence analysis (Figure 2). The recombinant protein expressed by pET vector had a FHV RNA2 fusion protein.
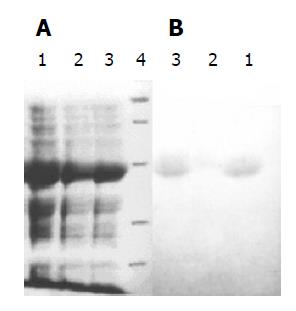
Three recombinant plasmids pETRNA2-I1E1, pETRNA2-I2E1, pETRNA2-I3E1, were carrying the HCV E1 epitope gene, expressed in E. coli BL21 (DE3). The yield of recombinant proteins was as high as 40% of the total cell proteins (Figure 4A). The recombinant proteins RNA-I1E1, RNA-I2E1, RNA-I3E1, were HCV E1 epitopes inserted in positions I1 (aa106), I2 (aa153), I3 (aa305) of the FHV capsid protein respectively. Better expression was found in pETRNA2-I1E1 and pETRNA2-I2E1.
Three recombinant proteins were separated by SDS-PAGE and transferred electrophoretically to nitrocellulose for immunoblotting as shown in Figure 4B. The three proteins showed immunoreactivity. RNA2-I1E1, RNA2-I3E1 had strong reactivity with HCV positive sera, and the reactivity of pETRNA2-I2E1 was relatively weaker.
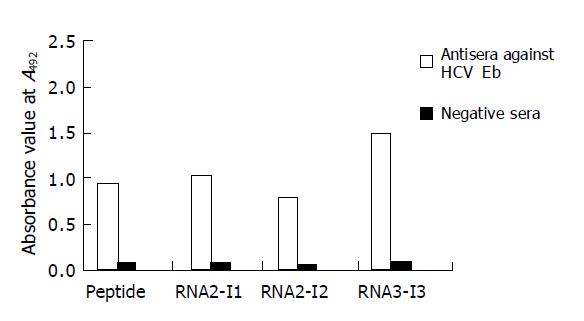
Three chimeric antigen proteins were further purified. After purification by Q-FF/HP negative ion-exchange and superdex 75 gel chromatography, the purity of these three chimeric proteins was higher than 90%. These chimeric antigens were coated respectively to microplates in 50 ng each well. Other wells were coated with synthetic HCV E1 peptide 5 μg/mL (500 ng/100 μL/well) in sodium carbonate buffer (pH 9.6) overnight. Antisera against HCV Eb and negative sera were used in ELISA. The results showed that the reactivities of two recombinant proteins (RNA2-I1E1, RNA2-I3E1) were greater than those of synthetic peptide (Figure 5).
It is well known that the immunogenicity of a peptide depends not only on its sequence, but also on the mode of presentation to the immune system. Attempts to increase the immunogenicity of these antigens used as vaccines have led to the development of a number of antigen presentation systems. Many of these are designed to present the antigen as a polyvalent particulate structure. Various types of particles have been used to present foreign epitopes: core antigen (HbcAg) and surface antigen (HBsAg) from HBV, capsid proteins from polio virus[18,19]. Using a human rhinovirus capsid sequence as a guest peptide and particles formed by HbcAg as a carrier, show that the internal location of the foreign sequence improves immunogenicity of that epitope by 10 to 50-fold when compared to the amino terminus location[20]. Furthermore, both constructs present the epitope more efficiently to the mAbs, than the free peptide. Since the properties of a given epitope might be influenced by its conformation, it should have a carrier system with multiple entry sites conferring many possible conformations. We developed a new protein carrier (FHV capsid protein) based on the structure known at atomic level for the location of foreign eptitopes.
Studies of the HCV antigenic structure have allowed construction of diagnostic systems with improved assay specificity and sensitivity[21,22]. HCV infection is generally detected using recombinant antigens from the conserved capsid, NS3 and NS4 regions[23], but the specificity and sensitivity of some regions are not sufficient. Recent studies have identified several epitopes in the envelope protein. Research in epitopes of envelope protein will help develop diagnostic reagents and recombinant vaccines for HCV. Since the nucleotide mutation rate in envelope E1 (35%) is higher than in core (8%) of HCV, we studied the expression and immunogenicity of a highly conserved B-cell epitope of E1 among different HCV genotypes. Reactivity of this epitope to synthetic peptide was observed in 10 of 11 sera from HCV-infected humans and 11 of 15 sera from chimpanzee[9].
The conserved B-cell epitope gene of HCV E1 was synthesized and inserted into the three portions of the RNA2 encoding the FHV capsid protein as the vector.
The three chimeric genes were cloned to prokaryotic expression vector pET-3 and expressed in E. coli BL21 (DE3). To improve the yield, we used the TB-P medium (the phosphate-rich medium).
All the hybrid proteins reacted with antiserum of HCV positive patients in Western blot test, but RNA2-I1E1 and RNA2-I3E1 had a stronger reactivity with HCV positive sera than pETRNA2-I2E1, suggesting that the same epitope inserted into different positions of FHV capsid protein surface may have different conformations. In this way, the inserted epitope is placed on the exterior surface of the protein. Therefore, the epitope is predominantly recognized by HCV-positive sera. These antigens are extremely useful in studying the influence of the stereochemistry of the presenting molecule in the recognition of a short heterologous amino acid sequence by human immune system.
The reactivity of RNA2-I1E1, RNA2-I3E1 is much higher than that of synthetic peptide, and the synthetic peptides are presented in 10-fold molar than recombinant proteins, suggesting that antigenicity of HCV E1 epitope can be greatly enhanced by inserting the epitope in positions of FHV capsid protein surface. The same results are observed using a human rhinovirus capsid sequence as a guest peptide and particles formed by HBcAg as a carrier. Both constructs (the epitopes inserted in two different positions) present the epitopes more efficiently to the mAbs than the free peptide[20], indicating that the region is highly immunogenic. This is probably due to the fact that the peptide in solution, unlike in the solid phase or the epitope inserted in the carrier protein, can exist in several metastable conformations[24], some of which are more likely to acquire the native structure.
The results obtained in this study suggest that the FHV capsomer system has the potential and flexibility for presentation of conformational epitopes.
Science Editor Wang XL Language Editor Elsevier HK
| 1. | Itoh K, Suzuki K, Ishiwata S, Tezuka T, Mizugaki M, Suzuki T. Application of a recombinant Fab fragment from a phage display library for sensitive detection of a target antigen by an inhibition ELISA system. J Immunol Methods. 1999;223:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 2. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4657] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 3. | Choo QL, Weiner AJ, Overby LR, Kuo G, Houghton M, Bradley DW. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull. 1990;46:423-441. [PubMed] |
| 4. | Ramsay ME, Balogun MA, Teo CG, Mortimer PP. Accuracy of perceptions of hepatitis B and C status. Injecting drug users need vaccination against hepatitis B. BMJ. 2000;320:512; author reply 513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 5. | López-Labrador FX, Ampurdanès S, Giménez-Barcons M, Guilera M, Costa J, Jiménez de Anta MT, Sánchez-Tapias JM, Rodés J, Sáiz JC. Relationship of the genomic complexity of hepatitis C virus with liver disease severity and response to interferon in patients with chronic HCV genotype 1b infection [correction of interferon]. Hepatology. 1999;29:897-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2343] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 7. | Miyamura T, Saito I, Katayama T, Kikuchi S, Tateda A, Houghton M, Choo QL, Kuo G. Detection of antibody against antigen expressed by molecularly cloned hepatitis C virus cDNA: application to diagnosis and blood screening for posttransfusion hepatitis. Proc Natl Acad Sci USA. 1990;87:983-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 118] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 8. | Saito M, Hasegawa A, Kashiwakuma T, Kohara M, Sugi M, Miki K, Yamamoto T, Mori H, Ohta Y, Tanaka E. Performance of an enzyme-linked immunosorbent assay system for antibodies to hepatitis C virus with two new antigens (c11/c7). Clin Chem. 1992;38:2434-2439. [PubMed] |
| 9. | Wang YF, Brotman B, Andrus L, Prince AM. Immune response to epitopes of hepatitis C virus (HCV) structural proteins in HCV-infected humans and chimpanzees. J Infect Dis. 1996;173:808-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Ishida C, Matsumoto K, Fukada K, Matsushita K, Shiraki H, Maeda Y. Detection of antibodies to hepatitis C virus (HCV) structural proteins in anti-HCV-positive sera by an enzyme-linked immunosorbent assay using synthetic peptides as antigens. J Clin Microbiol. 1993;31:936-940. [PubMed] |
| 11. | Ray R, Khanna A, Lagging LM, Meyer K, Choo QL, Ralston R, Houghton M, Becherer PR. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J Virol. 1994;68:4420-4426. [PubMed] |
| 12. | Okamoto H, Kurai K, Okada S, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 385] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 13. | Ching WM, Wychowski C, Beach MJ, Wang H, Davies CL, Carl M, Bradley DW, Alter HJ, Feinstone SM, Shih JW. Interaction of immune sera with synthetic peptides corresponding to the structural protein region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:3190-3194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Ferroni P, Mascolo G, Zaninetti M, Colzani D, Pregliasco F, Pirisi M, Barbone F, Gasparini V. Identification of four epitopes in hepatitis C virus core protein. J Clin Microbiol. 1993;31:1586-1591. [PubMed] |
| 15. | Moore JP, Cao Y, Conley AJ, Wyatt R, Robinson J, Gorny MK, Zolla-Pazner S, Ho DD, Koup RA. Studies with monoclonal antibodies to the V3 region of HIV-1 gp120 reveal limitations to the utility of solid-phase peptide binding assays. J Acquir Immune Defic Syndr. 1994;7:332-339. [PubMed] |
| 16. | Schutten M, Langedijk JP, Andeweg AC, Huisman RC, Meloen RH, Osterhaus AD. Characterization of a V3 domain-specific neutralizing human monoclonal antibody that preferentially recognizes non-syncytium-inducing human immunodeficiency virus type 1 strains. J Gen Virol. 1995;76:1665-1673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 17. | Scodeller EA, Tisminetzky SG, Porro F, Schiappacassi M, De Rossi A, Chiecco-Bianchi L, Baralle FE. A new epitope presenting system displays a HIV-1 V3 loop sequence and induces neutralizing antibodies. Vaccine. 1995;13:1233-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Clarke BE, Newton SE, Carroll AR, Francis MJ, Appleyard G, Syred AD, Highfield PE, Rowlands DJ, Brown F. Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature. 1987;330:381-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 246] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 19. | Burke KL, Dunn G, Ferguson M, Minor PD, Almond JW. Antigen chimaeras of poliovirus as potential new vaccines. Nature. 1988;332:81-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 136] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 20. | Brown AL, Francis MJ, Hastings GZ, Parry NR, Barnett PV, Rowlands DJ, Clarke BE. Foreign epitopes in immunodominant regions of hepatitis B core particles are highly immunogenic and conformationally restricted. Vaccine. 1991;9:595-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Krajden M. Molecular detection of hepatitis C virus: impact of detection methodology on clinical and laboratory correlations. Crit Rev Clin Lab Sci. 1995;32:41-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Roggendorf M, Lu M, Meisel H, Riffelmann M, Schreier E, Viazov S. Rational use of diagnostic tools in hepatitis C. J Hepatol. 1996;24:26-34. [PubMed] |
| 23. | Chien DY, Choo QL, Tabrizi A, Kuo C, McFarland J, Berger K, Lee C, Shuster JR, Nguyen T, Moyer DL. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc Natl Acad Sci USA. 1992;89:10011-10015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 78] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Moore JP. The reactivities of HIV-1+ human sera with solid-phase V3 loop peptides can be poor predictors of their reactivities with V3 loops on native gp120 molecules. AIDS Res Hum Retroviruses. 1993;9:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.1] [Reference Citation Analysis (0)] |