Published online May 14, 2005. doi: 10.3748/wjg.v11.i18.2811
Revised: July 18, 2003
Accepted: July 6, 2004
Published online: May 14, 2005
AIM: To find out key genes responsible for hepatocarc-inogenesis and to further understand the underlying molecular mechanism through investigating the differential gene expression between human normal liver tissue and hepatocellular carcinoma (HCC).
METHODS: DNA microarray was prepared by spotting PCR products of 1000 human genes including 445 novel genes, 540 known genes as well as 12 positive (housekeeping) and 3 negative controls (plant gene) onto treated glass slides. cDNA probes were prepared by labeling normal liver tissue mRNA and cancer liver tissue mRNA with Cy3-dUTP and Cy5-dUTP separately through reverse transcription. The arrays were hybridized against the cDNA probe and the fluorescent signals were scanned. The data obtained from repeated experiments were analyzed.
RESULTS: Among the 20 couple samples investigated (from cancerous liver tissue and normal liver tissue), 38 genes including 21 novel genes and 17 known genes exhibited different expressions.
CONCLUSION: cDNA microarray technique is powerful to identify candidate target genes that may play important roles in human carcinogenesis. Further analysis of the obtained genes is helpful to understand the molecular changes in HCC progression and ultimately may lead to the identification of new targets for HCC diagnosis and intervention.
- Citation: Mao HJ, Li HN, Zhou XM, Zhao JL, Wan DF. Monitoring microarray-based gene expression profile changes in hepatocellular carcinoma. World J Gastroenterol 2005; 11(18): 2811-2816
- URL: https://www.wjgnet.com/1007-9327/full/v11/i18/2811.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i18.2811
Hepatocellular carcinoma (HCC) is one of the most frequent human malignancies in the world. It is the fifth most common cancer with an estimated 437000 new cases diagnosed annually (5.4% of all new cancer cases). It ranks fourth in mortality rate, following lung, stomach and colon cancers. HCC is a leading cause of cancer-related deaths in adults in Asia and sub-Saharan Africa. Initiation and progression of HCC are thought to be due to an accumulation of genetic changes involving numerous genes[1]. Genes potentially involved in HCC include the p53 gene on chromosome 17p13.1[2-4]. However, it is likely that many of the genes involved in the initiation and progression of HCC are currently unknown.
Identification and evaluation of new molecular parameters are of utmost importance in cancer research and treatment. Genetic information of new genomic technologies and approaches is accumulated at a rapid pace. It is now possible to use cDNA array to identify candidate target genes that play important roles in human carcinogenesis[5]. To identify and monitor gene expression profile changes in HCC specimens may explore not only the cause of these pathological changes, but also provide the opportunity to identify novel targets for disease detection and intervention. In this paper, we constructed a cDNA microarray for liver tissue by selecting genes that were nonredundant and preferentially expressed in hepatocellular tissue. We described the assembly and utilization of liver cDNA microarray designed to assess the differences in gene expression between normal liver and tumor tissues.
The microarray consisted of 445 novel genes, 540 known genes as well as 12 positive (housekeeping) and 3 negative controls (plant gene) provided by Shanghai Cancer Institute. PCR products were used as universal primers. After completion of PCR, 5 μL of each reaction product was checked by agarose gel electrophoresis, purified and dissolved in 50% DMSO solution. These target genes were arrayed from 384-well microtiter plates onto silylated microscope slides (Cel Associate, Inc.) using PixSys 7500 spotting Robotics (Cartesian Technologies Inc.). Printed arrays were UV crosslinked (65 mJ/cm) and ready for use.
Total RNA was extracted from 20 couple cancerous liver tissues and normal liver tissues using TRIzol reagent. The amount of total RNA from each sample was measured by a spectrophotometer, and its quality was checked by agarose-gel electrophoresis. mRNA was purified using Oligo dT. The fluorescent cDNA probes were made through reverse transcription.
The probes from normal tissues were labeled with Cy3-dUTP, those from cancerous tissues with Cy5-dUTP. Reverse transcriptase was Superscript II RNase H- (GIBCO). Cy3-dUTP and Cy5-dUTP were purchased from Amersham. Each reverse transcription reaction contained 2.5 μg of mRNA. Following the reverse transcription reaction, samples were treated with 2.5 μL of 1 mol/L sodium hydroxide for 10 min at 37 °C, then neutralized by adding 2.5 μL of 1 mol/L Tris-HCl (pH 6.8) and 2.0 μL of 1 mol/L HCl. Probe mixtures were precipitated by ethanol and suspended in 20 μL of hybridization buffer (5×SSC+0.2% SDS).
The slides were boiled in water for 30 s to denature the cDNA, the probe mixtures were heated to 99 °C for 4 min, immediately chilled on ice for 3 min and applied to slides. The slides were covered with glass coverslips, hybridized overnight, incubated at 60 °C in a humid chamber. The slides were washed in 2×SSC+0.1% SDS for 5 min, 0.1×SSC+0.1% SDS for 5 min at room temperature. After a quick rinse in 0.1×SSC, the slides were air dried for scanning.
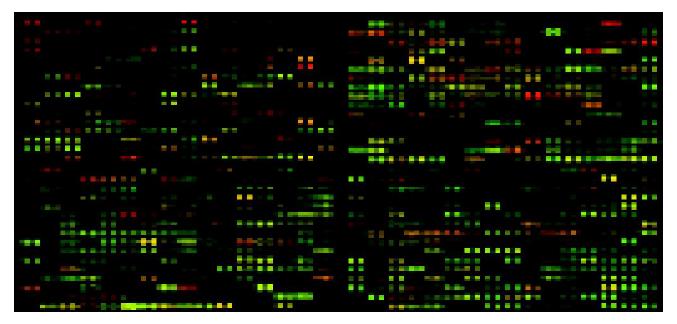
Microscope slides were scanned for Cy3 and Cy5 fluorescences using the Gene Pix4000B (Axon Instruments) at two wavelengths, and the obtained data (16-bit tiff image) were analyzed by GenPix Pro4.0 software, converting the signal intensity of each spot into a text format. The intensities of each spot at the two wavelengths represented the quantity of Cy3-dUTP and Cy5-dUTP respectively, and the overall intensities were normalized according to the ratios of the located 12 housekeeping genes. In each sample, the Cy3/Cy5 ratio values were log transformed, and global equalization to remove a deviation of the signal intensity between whole Cy3- and whole Cy5-fluorescences was performed by subtracting a median of all log(Cy3/Cy5) values from each log(Cy3/Cy5) value. Accurate differential expression measurement was obtained by taking the average of the ratios of three independent hybridizations. The processed overlay image of a hybridized slide is shown in Figure 1.
To investigate the gene expression profile changes in the development of HCC, a cDNA microarray was assembled with selected clones. The array consisted of a total of 1000 genes, including 115 novel genes, 120 unknown function genes with homology to cDNA sequences, 210 unknown function genes with homology to genomic sequences, 540 known genes as well as 12 positive (housekeeping) and 3 negative controls (plant gene). Each gene was spotted twice. Thus, there were 2000 spots on the array. The probes made from purified mRNA from HCC samples were labeled with the fluorescent dye Cy5 and mixed with the probes made from purified mRNA from normal liver tissues with the fluorescent dye Cy3.
In order to reduce the error occurrence in statistics, normal liver tissue mRNA was labeled separately with Cy3-dUTP and Cy5-dUTP as mixed probes. The array was hybridized independently thrice with the above mixed probes. The mean value from ratios of Cy5/Cy3 was applied to a normalization factor in further experiments. In addition, we noticed that these negative control spots showed a low intensity of signals after hybridization, which proved the reliability of statistics.
To investigate and monitor the gene expression profile changes in liver cancers, replicates of the fabricated cDNA array were hybridized independently with cDNA probes that were generated from 20 couples of different specimens of normal liver and cancer liver tissues (Figure 1). To examine the reliability of hybridization signals and experimental variables unrelated to the differences in hybridization probes, each couple specimens from normal liver and cancer liver tissues were hybridized twice independently. Forty groups of data were obtained. With each gene, variance among the cancer and normal samples was calculated.
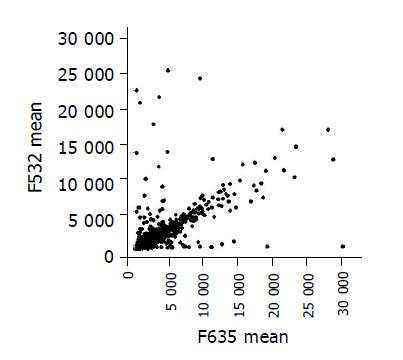
Scatter plots of the values of Cy3 and Cy5 fluorescent signals (Figure 2) also revealed a pattern of distribution and were clustered in a diagonal line as expected.
After the hybridization results were analyzed, 38 genes whose ratios between Cy5 signals and Cy3 signals were greater than 2 or less than 0.5 were selected for further analysis, including 21 novel genes (Table 2). Of these, 20 genes with a ratio between Cy5 and Cy3 signals greater than 2 showed a higher expression in cancerous tissues, 18 genes with a ratio between Cy5 and Cy3 signals less than 0.5 showed a lower expression in cancerous tissues. Seventeen of the 38 genes were known genes registered in GenBank (Table 1).
| Clone ID | GB-ACC | Type | Code | Identity | Remarks |
| 192 | Z11793 | Known-related | Redox | Selenoprotein | Related to redox |
| 273 | AF132609 | Known-related | DR | Histone deacetylase 6 | Related to DNA replication and transcription |
| 568 | Y07846 | Known-related | TS | GAR-22 meningioma deleted gene, 22q12 | Deleted in meningioma |
| 671 | X80199 | Known-related | O | MLN51 | Possibly related to breast cancer progression (17q11-q21.3) |
| 1483 | U50330 | Known-related | P | PCP-2 | Procollagen C-proteinase |
| 2648 | AJ223951 | Known-related | CC | CDK2 | CDK2 |
| 2803 | E12644 | Known-related | CC | Q6 (quiescin) | Cell proliferation inhibitor |
| 2820 | U89867 | Known-related | ST | Nuclear matrix protein 55 (nmt55), | Possibly related to ER negative phenotype and |
| related to breast cancer metastasis | tumor progression metastases in breast cancer | ||||
| 2939 | M25915 | Known-related | P | Complement cytolysis inhibitor, CLI. | Complement cytolysis inhibitor |
| 3120 | X86691 | Known-related | TA | Helicase (presumed), Mi-2 protein (218 ku), involved in transcriptional activation | |
| 3373 | U60068 | Known-unrelated | M/S | Fibronectin | Matrix protein fibronectin |
| 3423 | AF027740 | Known-related | Redox | Transferase, microsomal glutathione S-transferase 1-like 1 (MGST1L1) | Related to redox |
| 4367 | AF064770 | Known-related | ST | Kinase, DAG (diacylglycerol) kinase alpha | Kinase for diacylglycerol |
| 6655 | AF069987 | Known-related | TS | Nitrilase 1 (NIT1) | A putative tumor suppressor, with diadenosine triphosphate (A-ppp-A) hydrolase activity |
| 7072 | X57352 | Known-related | I | Interferon-inducible gene family (1-8 U gene) | |
| 8343 | X13905 | Known-related | ST | Ras-related rab1B protein | |
| 9039 | U41767 | Known-related | P | Metargidin | A membrane-anchored metalloprotease-disintegrin protein with an RGD integrin binding sequence |
| Clone ID | GB-ACC | Type | Identity |
| 6 | - | Novel | - |
| 50 | AC007078 | Novel-only DNA | Clone NH0340A14 |
| 432 | - | Novel | - |
| 775 | G42524 | Novel-only DNA | STS genomic (sequence tagged site) |
| 894 | - | Novel | - |
| 1008 | G27158 | Novel-only DNA | STS SHGC-31700 |
| 1485 | Z68162 | Novel-only DNA | Cosmid L145E5 |
| 1511 | AB002307 | Novel-unknown-function mRNA, | KIAA0309 |
| 1863 | AL110272 | Novel-unknown-function | DKFZp564C0716 |
| 2202 | AF086469 | Novel-unknown-function | Full-length insert cDNA clone ZD87A06 |
| 3227 | - | Novel | - |
| 3429 | AB020685 | Novel-unknown-function mRNA, | KIAA0878 |
| 4189 | - | Novel | - |
| 6068 | - | Novel | - |
| 7478 | AF052114 | Novel-only DNA | Clone 23887 |
| 7518 | - | Novel | - |
| 9427 | AB002339 | Novel-unknown-function mRNA, | KIAA0341 |
| 12723 | - | Novel | - |
| 13734 | AF030453 | Novel-only DNA | Chromosome 7q22, 7q22-q31.1 |
| 14214 | - | Novel | - |
| 14733 | - | Novel | - |
The green spots represent the genes that were more abundant in HCC than in normal tissues. The red spots represent the genes that were less abundant in HCC than in normal tissues. Yellow spots represent the genes that were equally expressed. Black spots represent the genes that were not expressed.
Our results indicated that the micro-array approach described here could identify differentially expressed novel genes. It might be the result of these novel genes obtained by screening the human normal liver cDNA library.
Among 20 couple samples investigated, mRNA profile analysis indicated that 38 genes including 21 novel genes and 17 known genes exhibited different expressions.
The 17 known genes cover a broad range of functional activities: transcription (TA), signaling transduction (ST), cell-cycle regulation (CC), protease/inhibitor (extracellular and cytoplasmic) metastasis related (P), redox regulator (energy related) (Redox), cancer (tumor) suppressor (TS), proto-oncogene (O), immune response related (I), DNA replication and repair (DR), matrix/stroma (M/S).
HCC is one of the most malignant tumors with a high mortality, and is characterized by aggressive growth behaviors and a high recurrence rate. During proliferation of a primary tumor or the establishment of metastatic foci, there was continuous remodeling of the extracellular matrix including various degrees of biosynthesis, reformation and degradation.
Some genes were overexpressed in HCC samples in our study. Metastatic lymph node 51 (MLN51) cDNA was isolated by screening a human breast cancer metastasis cDNA library. MLN51 cDNA encodes a novel human protein of 703 residues that shares no significant homology to any known protein. However, MLN51 is well conserved between vertebrate and invertebrate species suggesting an important biological function. The amino terminal half of the protein contains a coiled-coil domain and two potential nuclear localization signals. MLN51 is ubiquitously expressed in normal tissues. Human breast carcinomas show MLN51 overexpression in malignant epithelial cells. The uncommon association of protein-protein interaction domains often found either in nuclear or in cytoplasmic signaling proteins has raised a possible nucleo-cytoplasmic function for MLN51[6]. PCP-2 is a human receptor protein tyrosine phosphatase of the MAM domain family in human pancreatic adenocarcinoma cells. It is co-localized with beta-catenin and E-cadherin at cell junctions. Its intracellular part consists of two tandem phosphatase domains and a relatively large juxtamembrane region that is homologous to the conserved intracellular domain of cadherins, suggesting a role in the regulation of cell adhesion. PCP-2 is endogenously expressed at the cell surface and unregulated by increased cell density. PCP-2 is a negative regulator for cell migration. Interaction of PCP-2 with its substrate beta-catenin might be involved in the process of cell-cell contact[7]. Fibronectin (FN) is a glycoprotein component of connective tissue. It is involved in cancer progression. FN connects with cancer cells via its receptors, including integrins α5β1 and αvβ3. The ability in adhesion to FN and migration increased markedly, after HCC cells were transfected with H-ras oncogenes. In addition, FN has been reported to be the primary protein involved in the displacement of MMP-2 produced by adjacent normal cells to cancer tissues[8]. MMP-2 is associated with cell surface through its COOH-terminal and hemopexin-like domain via a number of mechanisms, including binding to cell-associated collagens IV and I. This may enhance the FN-induced displacement of MMP-2, and facilitate invasion of cancerous cells. IFN-inducible genes 1-8U that were overexpressed in cancer tissues may be cancer-related genes. IFN-inducible genes mediate several kinds of functional roles. IFN-inducible genes such as double-stranded RNA-dependent protein kinase could exert an antiviral and antiproliferative effects[9]. 1-8U expression in severely inflamed mucosae and colitis-associated cancer tissues in ulcerative colitis (UC) may also be important in protection against the proliferation of inflammation-mediated cells and tumor cells. The study indicated that expression of 1-8 U gene in colonic mucosa might be a useful marker in the identification of high-risk groups of UC-associated colon cancer[10].
In our study, we also found that some genes were down-regulated in cancer tissues. Selenium (Se) has been shown to prevent cancer in animal model systems when fed at levels exceeding the nutritional requirements[11,12]. A landmark study by Clark et al[13] showed cancer chemopreventive efficacy using a Se supplement in humans. The term “selenoprotein” is restricted to the proteins containing selenocysteine[14]. Selenoproteins such as mammalian thioredoxin reductase might play a role in Se-mediated cancer prevention[15]. A human 15 ku selenoprotein (Sep15) has been identified to contain selenium in the form of Sec in human T-cells and is present in various human tissues, such as liver, kidney, testis, brain, but the level of this selenoprotein was reduced substantially in a malignant prostate cell line and in HCC[16]. Quiescent Q6 (QSCN6) expression was induced just as fibroblasts began to leave the proliferating cycle and entered quiescence. QSCN6 is located on human chromosome 1q24, near the putative hereditary prostate cancer locus (HPC1). Expression of QSCN6 mRNA was suppressed in transformed fibroblasts. Quiescin QSCN6 potentially involves cell growth and the redox state of key proteins. The identification of QSCN6 as a gene that was highly expressed in quiescent cells compared to actively growing cells suggested that QSCN6 might play a role in the process by which normal cells enter a reversible quiescence. Inhibition of QSCN6 activity could thus play a role in cancer. The molecular characterization of the promoters of quiescence-induced genes and their molecular structures would shed new lights upon the mechanisms, which regulate the onset of quiescence and how these are deranged in cancer. nmt55/p54nrb may be critical to cell growth and function; decreased nmt55/p54nrb expression in ER- human breast tumors or the expression of nmt55/p54nrb variants in ER+ tumors may indicate loss of normal growth. nmt55/p54nrb has been shown in several studies to bind to RNA and interact with PSF and topoisomerase[17-21]. These results associated with several splicing factors essential for spliceosome formation (unpublished) and tumor hormonal status[22] suggest that nmt55/p54nrb may be involved in pre-mRNA processing. There is evidence for the role of RNA processing in oncogenesis[23,24]. The protein MGST1-L1, involved in redox regulation, has been reported to be under the control of p53[25]. The protein was also independently identified as an EST clone, and the full-length sequence was deposited in the GenBank database. The finding that MGST1-L1 was upregulated after p53 expression in a colorectal cancer cell line[25] suggests an important biological function possibly associated with cancer or apoptosis. These genes down-expressed in HCC in our study may play the same role.
These data indicated that we obtained the same results by using cDNA microarray by other methods and confirmed the feasibility, accuracy and effectiveness of microarray as a method to investigate the expression profiles of liver cancer.
The genes discussed here are those with a better-known function. However, the recognition of altered expression of genes with unknown function in Table 2 can help assign it a role, so these new genes will be subjected to further experiments to study their function.
In this paper, we demonstrated the utility of cDNA microarray in analyzing the molecular changes in HCC (Cy5) and normal liver (Cy3). Most importantly, the hybridization results might provide valuable information for further understanding the primary function of these new genes and molecular pathways or areas that may be involved in hepatic carcinogenesis. Monitoring and understanding the molecular changes in HCC progression will lead to the identification of new targets for HCC diagnosis and intervention in the future.
DNA microarrays have become an indispensable tool for the study of gene expression on a genomic scale[26,27]. At present, human cDNA collection remains the most completely representative and best characterized of all mammalian species. Human cDNA microarray is therefore the most powerful and convenient tool among all kinds of microarrays. We presented the methodology and results of using human cDNA microarray to study oncogenes and related genes. In addition, this method can contribute to analyzing multiple tumor samples in a rapid fashion. Using cDNA microarrays to define alterations in gene expression associated with a specific cancer may be an efficient way to uncover the clues to specific molecular derangements that account for its pathogenesis and thus identifying potential targets for therapeutic intervention. Moreover, recognition of pathognomonic alterations in gene expression might provide a basis for improved diagnosis and therefore allowing selection of the most appropriate therapeutic strategies.
| 1. | Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H. Allelotype study of primary hepatocellular carcinoma. Cancer Res. 1991;51:89-93. [PubMed] |
| 2. | Wang XW, Hussain SP, Huo TI, Wu CG, Forgues M, Hofseth LJ, Brechot C, Harris CC. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181-182:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 145] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 3. | Okada T, Iizuka N, Yamada-Okabe H, Mori N, Tamesa T, Takemoto N, Tangoku A, Hamada K, Nakayama H, Miyamoto T. Gene expression profile linked to p53 status in hepatitis C virus-related hepatocellular carcinoma. FEBS Lett. 2003;555:583-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat Genet. 1995;11:447-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 258] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol. 1999;154:981-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 267] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 6. | Degot S, Régnier CH, Wendling C, Chenard MP, Rio MC, Tomasetto C. Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene. 2002;21:4422-4434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Yan HX, He YQ, Dong H, Zhang P, Zeng JZ, Cao HF, Wu MC, Wang HY. Physical and functional interaction between receptor-like protein tyrosine phosphatase PCP-2 and beta-catenin. Biochemistry. 2002;41:15854-15860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Saad S, Gottlieb DJ, Bradstock KF, Overall CM, Bendall LJ. Cancer cell-associated fibronectin induces release of matrix metalloproteinase-2 from normal fibroblasts. Cancer Res. 2002;62:283-289. [PubMed] |
| 9. | Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 441] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 10. | Horiuchi K, Weskamp G, Lum L, Hammes HP, Cai H, Brodie TA, Ludwig T, Chiusaroli R, Baron R, Preissner KT. Potential role for ADAM15 in pathological neovascularization in mice. Mol Cell Biol. 2003;23:5614-5624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 143] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 11. | Combs GF, Gray WP. Chemopreventive agents: selenium. Pharmacol Ther. 1998;79:179-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 424] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 12. | Ip C. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128:1845-1854. [PubMed] |
| 13. | Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276:1957-1963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1666] [Cited by in RCA: 1343] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 14. | Mostert V. Selenoprotein P: properties, functions, and regulation. Arch Biochem Biophys. 2000;376:433-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 78] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis. 1999;20:1657-1666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 463] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 16. | Coppock DL, Cina-Poppe D, Gilleran S. The quiescin Q6 gene (QSCN6) is a fusion of two ancient gene families: thioredoxin and ERV1. Genomics. 1998;54:460-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356-3367. [PubMed] |
| 18. | Rossi F, Labourier E, Forné T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 249] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 19. | Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273:26261-26264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Straub T, Knudsen BR, Boege F. PSF/p54(nrb) stimulates "jumping" of DNA topoisomerase I between separate DNA helices. Biochemistry. 2000;39:7552-7558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Basu A, Dong B, Krainer AR, Howe CC. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol Cell Biol. 1997;17:677-686. [PubMed] |
| 22. | Traish AM, Huang YH, Ashba J, Pronovost M, Pavao M, McAneny DB, Moreland RB. Loss of expression of a 55 kDa nuclear protein (nmt55) in estrogen receptor-negative human breast cancer. Diagn Mol Pathol. 1997;6:209-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761-27764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 178] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 24. | Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233-2239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 251] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 25. | Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1866] [Cited by in RCA: 1878] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 26. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5103] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 27. | Khan J, Bittner ML, Chen Y, Meltzer PS, Trent JM. DNA microarray technology: the anticipated impact on the study of human disease. Biochim Biophys Acta. 1999;1423:M17-M28. [PubMed] |