Published online May 14, 2005. doi: 10.3748/wjg.v11.i18.2704
Revised: April 5, 2004
Accepted: May 24, 2004
Published online: May 14, 2005
AIM: To produce high-quality polyclonal antibody to lysosome-associated protein transmembrane 4B-35 and to identify LAPTM4B-35 expression in cancer tissues and its correlation with differentiation status of hepatocellular carcinoma (HCC).
METHODS: The 297 bp 5’ end of LAPTM4B cDNA was obtained by PCR and inserted into prokaryotic expression vector pGEX-KG. Then the recombinant pGEX-KG-N1-99 was transformed into E.coli JM109 to express GST-fusion protein. The fusion protein was purified by glutathione sepharoseTM 4B agarose. The purified GST-LAPTM4B-N1-99 was characterized by SDS-PAGE, and used to immunize rabbits. The titer and specificity of antisera were detected by ELISA and Western blot, respectively. The correlation between the expression levels of LAPTM4B-35 and the differentiation status of HCC was analyzed via Western blot. The expression of LAPTM4B-35 in HCC and other six cancer tissues was investigated via tissue chip and immunohistochemical analysis.
RESULTS: About 6.2 mg of pure GST-LAPTM4B-N1-99 was isolated from 1 L of bacteria. The GST-LAPTM4B-N1-99 produced high titer antisera in rabbits and showed good immunity. Western blot showed specific reactions for the antibody to the LAPTM4B-35 in the total proteins from HCC tissues and BEL-7402 cells, also to the fusion protein purified or in the transformed bacteria. LAPTM4B-35 was remarkably expressed in several cancers, such as HCC, breast cancer, gastric carcinoma, lung cancer, and colon carcinoma, but not commonly expressed in esophageal cancer and rectum carcinoma. Notably, the expression levels of LAPTM4B-35 were significantly and inversely correlated to the differentiation of HCCs in a 20 case analysis.
CONCLUSION: Specific polyclonal antibody (LAPTM4B-N1-99-pAb) to LAPTM4B-35 was produced. It identified the expression of LAPTM4B-35 in some cancer tissues originated from single layer cuboidal and columnar epithelial cells and firmly demonstrated that the expression of LAPTM4B-35 in HCC was inversely correlated with the differentiation of HCC.
- Citation: Peng C, Zhou RL, Shao GZ, Rui JA, Wang SB, Lin M, Zhang S, Gao ZF. Expression of lysosome-associated protein transmembrane 4B-35 in cancer and its correlation with the differentiation status of hepatocellular carcinoma. World J Gastroenterol 2005; 11(18): 2704-2708
- URL: https://www.wjgnet.com/1007-9327/full/v11/i18/2704.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i18.2704
Hepatocellular carcinoma (HCC) remains one of the commonest internal malignancies of mankind. It ranks fifth in frequency in the world with an estimated number of 0.5-1 million cases per year, most of which occur in sub-Saharan Africa and south-east Asia including China[1-3]. The vast majority can be attributed to chronic hepatitis B virus (HBV) infection and there has been almost no change in incidence. It has also become evident that the number of HCC is increasing in low to middle-incidence countries such as USA[4-6], France[7,8], UK[9,10] and Japan[11,12]. The rising number of cases in these countries is most likely to be due to chronic hepatitis C virus (HCV) infection[4,6,12].
LAPTM4B, which was first cloned by our research group as a novel oncogene candidate, is highly expressed in vast majority of HCCs[13,14]. BLAST program analysis showed that the LAPTM4B was mapped to chromosome 8q22.1, the gain region in HCC shown by CGH analysis. The ORF of LAPTM4B contains two ATGs, indicating that this gene may encode two putative proteins with 35 kDa (317 aa) and 24 kDa (226 aa)[14,15], respectively. The former study confirmed that the LAPTM4B-35 was translated from the whole ORF and initiated from the first ATG, whereas the LAPTM4B-24 was translated from the second ATG[14]. The integral membrane protein characteristics were indicated by computer analysis and demonstrated by experiments. The LAPTM4B gene was widely expressed in normal human tissues shown by Northern blot[14]. Its expression was high in heart, skeletal muscle and testis; moderate in ovary, kidney; and pancreas; low in liver, spleen, and thymus; but lowest in lung and peripheral leukocytes. It was remarkably overexpressed (48 over 55 cases) in HCC when compared with PNL. Furthermore, the expression levels of LAPTM4B mRNA were significantly related to the differentiation status of HCCs: the highest in poorly-differentiated HCCs, higher in moderately-differentiated HCCs, and low in well-differentiated HCCs[14].
The biological effects of LAPTM4B were studied by transient and stable transfection. The result showed that cell proliferation was promoted via LAPTM4B stable transfection of both mouse NIH3T3 cells[16] and human HLE cells (manuscripts in preparation). Also, the LAPTM4B transfected NIH3T3 cells were tumorigenic when the transfectants were inoculated into NIH mice. Coimmunoprecipitation assay indicated that LAPTM4B interacted with integrin-α6β1 in BEL-7402 cells, which were enhanced by LN-1[15], and might play an important role in the integrin-α6 mediating signal transduction pathways. It was also found that the sequences of 91 amino acids at the N-terminus of LAPTM4B-35 were essential for its functions on cell survival and growth, which was revealed via transient transfection of plasmids containing full length and truncated sequences (273 bp) at the 5’ end of LAPTM4B ORF into HLE cells[14]. After 2-3 wk of G418 selection, colonies in pCDNA3-BE (containing truncated ORF) transfected cells were almost completely disappeared, whereas the pCDNA3-AE (containing full ORF) transfected cells formed lots of colonies[14]. These results indicate that LAPTM4B-35 plays an important role in the regulation of cell survival, proliferation, and may involve in carcinogenesis.
It was evidenced that the overexpression of LAPTM4B-35 promoted malignant transformation of some cell lines, including accelerated proliferation, migration and invasion of cells, and activated some protooncogenes, including immediate early genes, such as c-myc, c-fos and c-jun (manuscripts preparation).
To investigate the function and expression of LAPTM4B-35 in HCC and HCC cell lines, specific antibody to LAPTM4B-35, but not LAPTM4B-24, the 297 bp at 5’ end of LAPTM4B cDNA encoding LAPTM4B-N1-99 was cloned into donor vector pGEX-KG[17-19] and the recombinant plasmid was transformed into competent E. coli cells JM109. The GST-LAPTM4B-N1-99 fusion protein was produced in JM109 cells after induced with IPTG, and purified using glutathione sepharoseTM 4B agarose[20,21]. After rabbits were immunized, specific polyclonal antibodies, LAPTM4B-N1-99-pAb, against the N-terminus of LAPTM4B-35 were obtained. With this antibody, the expressions of LAPTM4B-35 in HCC and several cancer tissues were performed via Western blot and TMA[21-24], respectively. The correlation between expression levels of LAPTM4B-35 and HCC differentiation status were analyzed.
A total of 20 pairs of specimens were obtained from patients with HCC (aged from 35 to 70 years) who underwent hepatectomy at the Peking Union Medical College Hospital (the West Hospital) in Beijing. Histopathological analyses were independently performed by pathologists. Tumor differentiation was graded as I-III, according to the Edmon-dson Grading System[25,26]. Specimens were frozen immediately after surgical resection and stored in liquid nitrogen.
Molecular biological enzymes were purchased from Sangon Biotechnology Ltd. Peroxidase-conjugated goat anti-rabbit IgG (H+L) was purchased from Zhongshan Biotechnology Ltd. Glutathione-sepharose 4B agarose was purchased from Pharmacia Biotech Ltd.
A TMA slide was purchased from Chengdu Phargentech Ltd. It contained 60 species of tissues, including two normal esophageal epithelial and eight esophageal cancer tissues, two normal mammary gland and eight breast cancer tissues, two normal lung and eight lung cancer tissues, two normal gastric epithelial and eight gastric carcinoma tissues, two normal colonic epithelial and eight colon carcinoma tissues, two normal rectal epithelial and eight rectum carcinoma tissues. All specimens were embedded in paraffin.
To clone the 5’ end of LAPTM4B ORF, PCR method was used. Based on the published LAPTM4B gene sequence[14], two oligonucleotide primers (P1: 5’ GGGATCCGCCACCA-TGACGTCACGGACTCGG 3’; P2: 5’ GCGAAGCTTC-GTCCAGGGCGCGACCATC 3’) were synthesized (Sangon Biotechnology Ltd). P1 was extended to the 5’ end containing recognition sequences for endonuclease BamHI (underlined). P2 was extended to the 5’ end containing recognition sequences for endonuclease Hind III(underlined). PCR amplification was performed for 30 cycles at 94 °C for 30 s, at 55 °C for 30 s and at 72 °C for 50 s.
The pGEX-KG vector was digested with BamHI and Hind III. cDNA fragment at the 5’ end of LAPTM4B ORF was subcloned into the corresponding site of pGEX-KG to obtain a plasmid pGEX-KG-N1-99. Then the recombinant plasmid was transformed into the E.coli strain JM109. The transformed colonies containing the cloned cDNA were screened by restriction enzyme analysis and DNA sequencing.
An overnight LB culture of E.coli containing pGEX-KG-N1-99 was inoculated at a dilution of 1:100, and incubated at 37 °C with shaking. When the absorbance at 600 nm reached 0.6, GST-LAPTM4B-N1-99 expression was induced by the addition of 0.5 mmol/L IPTG. Further 4 h for growing, the E.coli strains were harvested by spinning at 4000 r/min for 15 min at 4 °C. The pellet was resuspended in PBS containing 0.05% Tween-20 of the original volume and lysed by sonication on ice, and then centrifuged at 10000 g for 20 min at 4 °C. The supernatant was passed through glutathione-sepharose 4 B beads according to the manufacturer’s instructions. SDS-PAGE was performed to analyze the purified protein from the infected cells.
Two rabbits were used to generate polyclonal antibodies against the fusion protein. Each rabbit was injected with 1 mg purified fusion protein in complete adjuvant in the initial injection and with uncompleted adjuvant for the following injection. Before injection, the rabbit blood was collected for producing pre-immune serum. The titer of antisera was measured by ELISA.
Proteins in cell lysates and tissues were fractionated by 10% SDS-PAGE and followed by electrotransferring onto nitrocellulose filters (Bio-Rad). The filters were blocked at 4 °C overnight with a blocking buffer (pH 7.6) containing 5% non-fat dry milk. Then the filters were incubated with indicated antibody (1:300) for 2 h at room temperature. After being washed with TBST (pH 6.0), the filters were incubated at room temperature for 1 h with horseradish peroxidase conjugated goat anti-rabbit IgG at 1:2000 dilution in TBS-0.5% non-fat dry milk. Immunoreactive bands were visualized using ECL detection reagents (Santa Cruz).
HCC tissue and TMA slide were fixed in 4% PFA in PBS for 10 min, washed twice with PBS, and incubated in 3% H2O2 to eliminate the endogenous peroxidase activities. The tissues were then incubated either at room temperature for 1 h or at 4 °C overnight with 1% goat serum in PBS to block the nonspecific binding of antibodies. The slides were further incubated sequentially with polyclonal antibody and goat anti-mouse IgG conjugated to horseradish peroxidase. The pre-immune serum was used as a control. Color developments were performed by incubation with 3,3’-diaminobenzidine tetrahydrochloride in 0.03% H2O2 and 50 mmol/L Tris-HCl, pH 7.4. Hematoxylin was used for counterstaining.
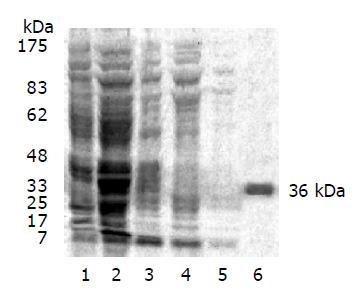
Confirmed by restriction endonuclease digestion and DNA sequencing, pGEX-KG-N1-99 plasmid was successfully constructed and GST-LAPTM4B-N1-99 was expressed in E.coli JM109 by IPTG induction. A 36 kDa fusion protein was stained in high intensity by 10% SDS-PAGE (Figure 1). This fusion protein was majored as a soluble pattern.
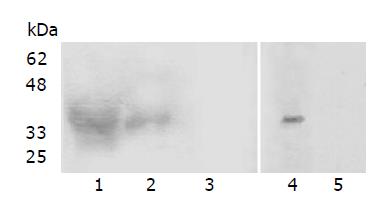
After being immunized thrice with purified GST-LAPTM4B-N1-99, two rabbits both generated antisera at a titer of 5×104, measured by ELISA. Western blot analysis showed that there was a specific Ag-Ab binding band at 36 kDa in the unpurified and purified GST-LAPTM4B-N1-99 (Figure 2A, lanes 1 and 2). The BEL-7402 cell lysate was also identified by LAPTM4B-N1-99-pAb (Figure 2B, lane 4).
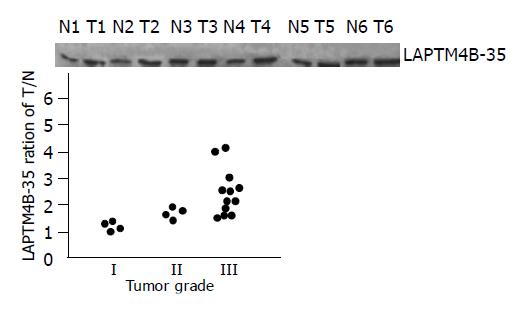
The expression levels of LAPTM4B-35 were significantly related to the differentiation status of HCCs: highest in poorly-differentiated HCCs (grade III, 12/20), high in moderately-differentiated HCCs (grade II, 4/20), and low in well-differentiated HCCs (grade I, 4/20) (Figure 3).
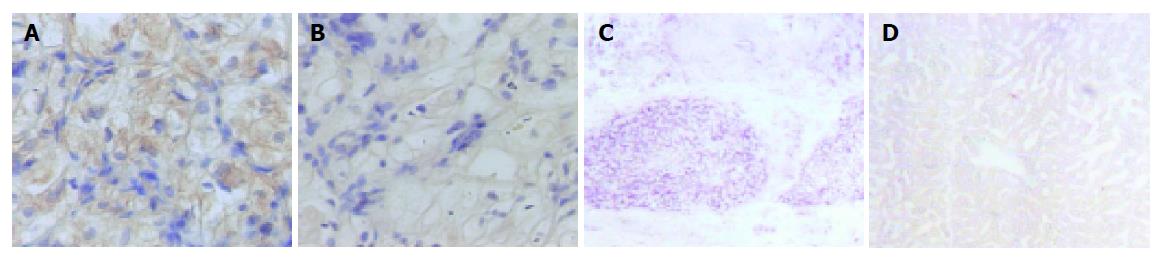
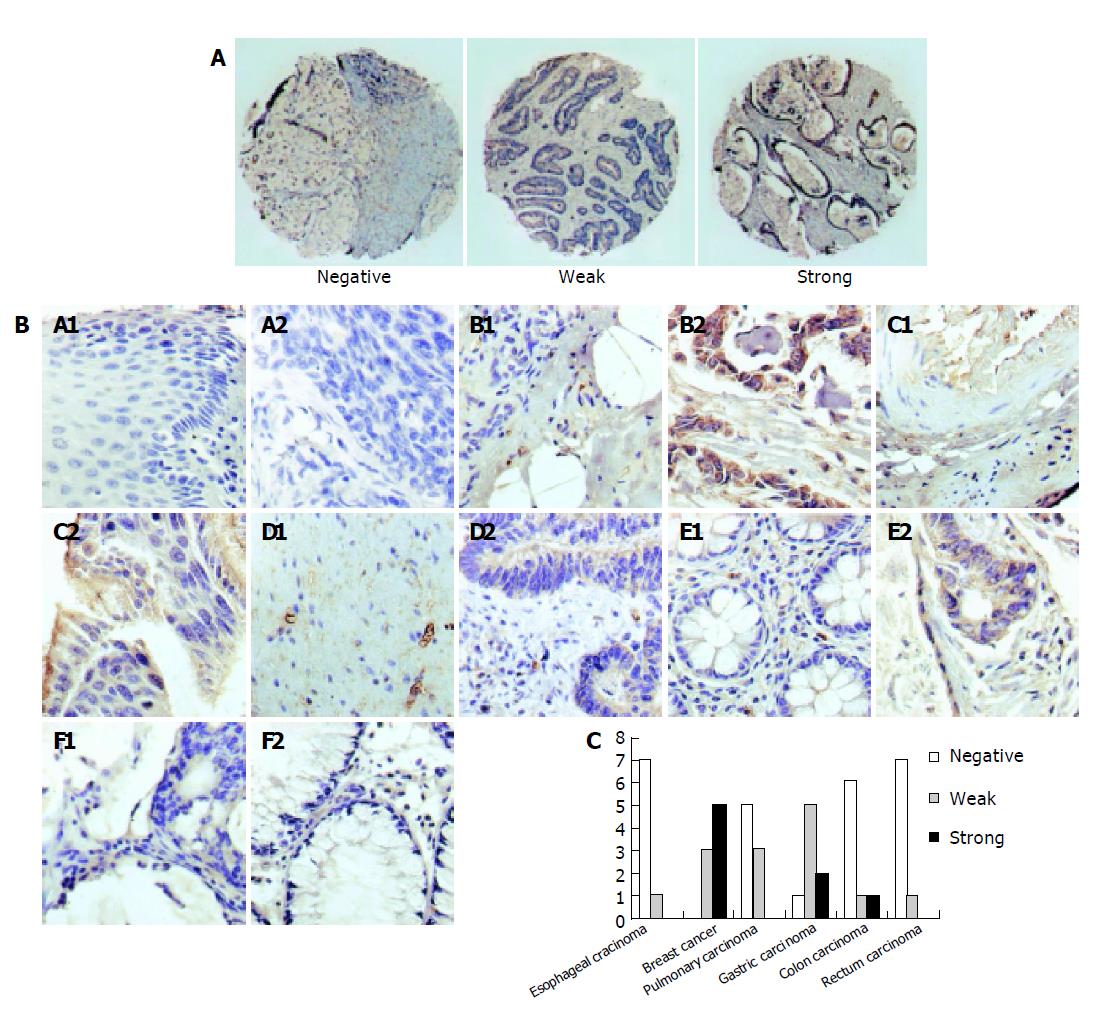
Immunohistochemical analysis was undertaken to characterize the expression of LAPTM4B-35 in HCC and TMA. In HCC (Figure 4A), positive brown signals were seen only in HCC cells and negative signals in the interstitial tissue of liver. The result was well coincident with the mRNA detection via in situ hybridization (Figure 4C)[14]. Moreover, the expression of LAPTM4B-35 was also analyzed via TMA in six types of cancer tissues: esophageal carcinoma, breast cancer, pulmonary carcinoma, gastric carcinoma, colon carcinoma, and rectum carcinoma. There were no significant LAPTM4B-35 expressions in six counterparts of normal tissues but obvious high and weak expression accounted for the majority of breast cancers and gastric carcinomas. Positive staining could only be observed in minority of pulmonary carcinomas and colon carcinomas and almost negative staining in all of esophageal carcinomas and rectum carcinomas tested (Figure 5).
In our previous work, we have produced a polyclonal antibody: LAPTM4B-EC2-pAb, which was prepared by immunization with KLH-conjugated 10-peptides whose sequences are localized at the second extracellular loop between the third and fourth transmembrane regions. By Western blot, expressions of LAPTM4B-35 and LAPTM4B-24 were identified in HCC, PNL and NL tissues with LAPTM4B-EC2-pAb, indicating that LAPTM4B-35 may initiate from the first ATG, whereas LAPTM4B-24 is translated from the second ATG. Notably, the expression levels of LAPTM4B-35 were primarily shown by Western blot with LAPTM4B-EC2-pAb to correlate with the differentiation status of 12 cases of HCC tissues, but the expression levels of LAPTM4B-24 did not relate to the differentiation status of HCC tissues.
To further clarify the correlation between upregulation of LAPTM4B-35 and differentiation of HCC and the function of LAPTM4B-35, we generated a specific antibody: LAPTM4B-N1-99-pAb, against LAPTM4B-35 but not against LAPTM4B-24. Applying this antibody, it was firmly demonstrated that the expression of LAPTM4B-35 protein was inversely related with the status of differentiation status of HCC, suggesting its importance in carcinogenesis and progress of HCC as well as a possibility of being a criterion of HCC differentiation and pathological grading. The expression levels of LAPTM4B-35 were not found to correlate with the AFP levels of sera.
Using LAPTM4B-N1-99-pAb, the expression of LAPTM4B-35 in some cancers originated from single layer cuboidal and columnar epithelia, such as HCC, breast cancer, gastric cancer and pulmonary cancer. We have previously reported that the high metastatic cell lines, such as the giant cell pulmonary carcinoma and prostate carcinoma cell lines highly expressed LAPTM4B-35. However, cancers originated from stratified epithelia, such as esophageal carcinoma and rectum carcinoma, did not practically express LAPTM4B-35. Malignant tumors from mesenchymal cells did not express LAPTM4B-35 either. The result of TMA gave us a clue that LAPTM4B-35 might also play an important role in breast cancer, gastric cancer or pulmonary carcinoma.
In conclusion, specific polyclonal antibody to the cytoplasmic N-terminal tail of LAPTM4B-35 was obtained and used for Western blot and immunohistochemistry. The expression of LAPTM4B-35 is associated with the differentiation status of HCC. In addition, the expression of LAPTM4B-35 in other cancer tissues may pave the way for further study on the function of N-terminal of LAPTM4B-35 for the development of tumors and their clinical diagnosis.
| 1. | Parkin DM, Muir CS. Cancer Incidence in Five Continents. Comparability and quality of data. IARC Sci Publ. 1992;45-173. [PubMed] |
| 2. | Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 671] [Cited by in RCA: 671] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 3. | Okuda K. Hepatocellular carcinoma. J Hepatol. 2000;32:225-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 398] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2221] [Cited by in RCA: 2140] [Article Influence: 82.3] [Reference Citation Analysis (0)] |
| 5. | Kaczynski J, Odén A. The rising incidence of hepatocellular carcinoma. N Engl J Med. 1999;341:451; author reply 452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 6. | Ince N, Wands JR. The increasing incidence of hepatocellular carcinoma. N Engl J Med. 1999;340:798-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 109] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Benhamiche AM, Faivre C, Minello A, Clinard F, Mitry E, Hillon P, Faivre J. Time trends and age-period-cohort effects on the incidence of primary liver cancer in a well-defined French population: 1976-1995. J Hepatol. 1998;29:802-806. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Deuffic S, Buffat L, Poynard T, Valleron AJ. Modeling the hepatitis C virus epidemic in France. Hepatology. 1999;29:1596-1601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 148] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 9. | Mant JW, Vessey MP. Trends in mortality from primary liver cancer in England and Wales 1975-92: influence of oral contraceptives. Br J Cancer. 1995;72:800-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Taylor-Robinson SD, Foster GR, Arora S, Hargreaves S, Thomas HC. Increase in primary liver cancer in the UK, 1979-94. Lancet. 1997;350:1142-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 370] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 11. | Okuda K, Fujimoto I, Hanai A, Urano Y. Changing incidence of hepatocellular carcinoma in Japan. Cancer Res. 1987;47:4967-4972. [PubMed] |
| 12. | Makimoto K, Higuchi S. Alcohol consumption as a major risk factor for the rise in liver cancer mortality rates in Japanese men. Int J Epidemiol. 1999;28:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Liu J, Zhou R, Zhang N, Rui J, Jin C. Biological function of a novel gene overexpressed in human hepatocellular carcinoma. Chin Med J (Engl). 2000;113:881-885. [PubMed] |
| 14. | Shao GZ, Zhou RL, Zhang QY, Zhang Y, Liu JJ, Rui JA, Wei X, Ye DX. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene. 2003;22:5060-5069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 15. | Liu X, Zhou R, Zhang Q, Zhang Y, Shao G, Jin Y, Zhang S, Lin M, Rui J, Ye D. Identification and characterization of LAPTM4B encoded by a human hepatocellular carcinoma-associated novel gene. Beijing DaXue XueBao. 2003;35:340-347. [PubMed] |
| 16. | He J, Shao G, Zhou R. Effects of the novel gene, LAPTM4B, highly expression in hepatocellular carcinoma on cell proliferation and tumorigenesis of NIH3T3 cells. Beijing DaXue XueBao. 2003;35:348-352. [PubMed] |
| 17. | Hakes DJ, Dixon JE. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem. 1992;202:293-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 207] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1398] [Cited by in RCA: 1534] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 19. | Ball EH, Shephard LB, Gill GN. Purification and properties of thyroid hormone receptor beta 1 expressed in Escherichia coli as a fusion protein. Protein Expr Purif. 1995;6:33-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 20. | Corsaro A, Thellung S, Russo C, Villa V, Arena S, D'Adamo MC, Paludi D, Rossi Principe D, Damonte G, Benatti U. Expression in E. coli and purification of recombinant fragments of wild type and mutant human prion protein. Neurochem Int. 2002;41:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Mercado-Pimentel ME, Jordan NC, Aisemberg GO. Affinity purification of GST fusion proteins for immunohistochemical studies of gene expression. Protein Expr Purif. 2002;26:260-265. [PubMed] |
| 22. | Hewitt SM. Design, construction, and use of tissue microarrays. Methods Mol Biol. 2004;264:61-72. [PubMed] |
| 23. | Zhang DH, Salto-Tellez M, Chiu LL, Shen L, Koay ES. Tissue microarray study for classification of breast tumours. Ann Acad Med Singapore. 2003;32:S75-S76. [PubMed] |
| 24. | Otsuka M, Kato M, Yoshikawa T, Chen H, Brown EJ, Masuho Y, Omata M, Seki N. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem Biophys Res Commun. 2001;289:876-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 66] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Kenmochi K, Sugihara S, Kojiro M. Relationship of histologic grade of hepatocellular carcinoma (HCC) to tumor size, and demonstration of tumor cells of multiple different grades in single small HCC. Liver. 1987;7:18-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 160] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Sasaki Y, Imaoka S, Ishiguro S, Nakano H, Kasugai H, Fujita M, Inoue E, Ishikawa O, Furukawa H, Nakamori S. Clinical features of small hepatocellular carcinomas as assessed by histologic grades. Surgery. 1996;119:252-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |