Published online May 7, 2005. doi: 10.3748/wjg.v11.i17.2647
Revised: April 11, 2004
Accepted: May 24, 2004
Published online: May 7, 2005
AIM: To assess the variability of adhesin gene hpaA between different Helicobacter pylori (H pylori) strains with PCR-restriction fragment length polymorphism (RFLP).
METHODS: Twelve different H pylori strains were chosen to amplify the 710-bp segments of gene hpaA. These strains were NCTC11637, SS1; Chongqing clinical isolates CCS9801, CCS9802, CCS9803, CCS9806, CCS9809, CCS9810, CCS9813, which were gained from patients of gastritis; Mongolia gerbil adapted H pylori strains (abbreviation MG), which were gained from the following steps: gastric mucosal specimens of Mongolia gerbils infected by clinical isolate CCS9803 were cultured and detected, the positive H pylori strains were named as the first generation of Mongolia gerbil adapted H pylori strains (abbreviation MG1) and then were subcultured with healthy Mongolia gerbil to generate MG2, in turn to gain the ninth generation (abbreviation MG9). All hpaA segments, obtained from 12 different H pylori strains, were digested by HhaI and HaeIII individually and analyzed by agarose gel electrophoresis.
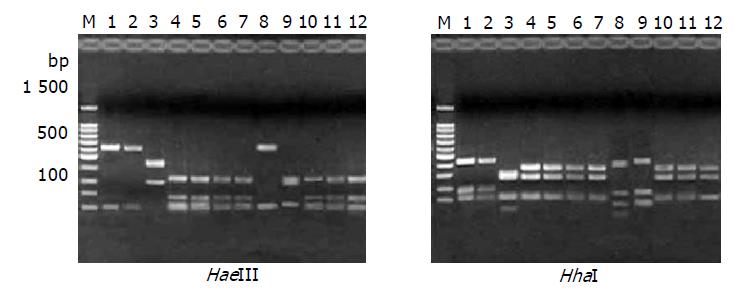
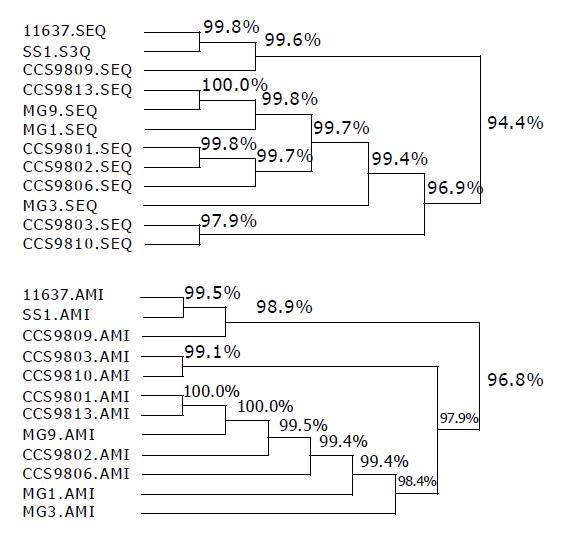
RESULTS: In all 12 strains, the 710-bp PCR products were successfully amplified and products were cloned to pMD18-T vector respectively, then the recombinant plasmids were digested simultaneously with NcoI and XhoI to recover the small fragments. The objective fragments from 12 different H pylori strains digested with Hae III could be seen as 4 types of bands and 5 types with Hha I. According to the hpaA RFLP patterns, the 12 H pylori strains could be divided into 5 groups: group I, NCTC11637 and SS1; group II, CCS9809, which RFLP type digested with HaeIII was the same as strains of group I, but HhaI RFLP showed difference compared with the other groups; group III, CCS9810; group IV, CCS9803; group V: CCS9801, CCS9802, CCS9806, CCS9813, MG1, MG3 and MG9. The sequence data of 12 hpaA segments were analyzed by DNAsis software and it was observed that: (1) The homologies of base pair and amino acid sequence between strains NCTC11637, SS1, CCS9809 were 99.6% and 98.9%, respectively; (2) The homology of base pair and amino acid sequence between CCS9803 and CCS9810 was 97.7% and 99.1%; (3) That of the rest strains, CCS9801, CCS9802, CCS9806, CCS9813, MG1, MG3, MG9 reached 99.4% and 98.4%; (4) The base pair homologies between all hpaA fragments of different sources were higher than 94.6%, therefore the correspondence of deduced amino acid sequence was higher than 96.8% between each other.
CONCLUSION: The gene hpaA from different H pylori strains revealed variation, and this might provide an effective method for molecular epidemiological survey of H pylori.
- Citation: Hong Y, Mao XH, Zeng WK, Ma LM, Jing SR, Zou QM. Restriction fragment length polymorphism of adhesin gene hpaA from different Helicobacter pylori strains of Chongqing, China. World J Gastroenterol 2005; 11(17): 2647-2652
- URL: https://www.wjgnet.com/1007-9327/full/v11/i17/2647.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i17.2647
Helicobacter pylori (H pylori) is a stomach-colonizing bacterium that not only causes chronic gastritis and duodenal ulcer disease but also is a risk factor for gastric cancer[1,2]. However, its source of infection, mode of transmission and etiology of recurrence have not been fully clarified. Because the phenotype characters of H pylori showed similarity, it could not be identified by traditional classification system. With the development of molecular biology and the application of new molecular biological technology, classification and epidemiological study of bacteria rely more and more on genetic characters[3], such as plasmid profiles, chromosomal restriction endonuclease analysis, ribotyping, PCR-restriction fragment length polymo-rphism (RFLP), PCR-SSCP, etc. Using these methods, we could differentiate H pylori strains, compare the similarity of different H pylori, track the source of H pylori infection, investigate the route of transmission, confirm the relapse to be recrudescence of the same strain or reinfection of other H pylori strains, and also carry out molecular epidemiological survey, explore distributing status and epidemic characteristics of H pylori[4-6].
In the present study, 710-bp gene hpaA from 12 H pylori strains were cloned and sequenced. Restriction endonucleases HhaI and HaeIII were chosen to do RFLP analysis.
Twelve different H pylori strains were NCTC11637, SS1 (kindly presented by China Microbiology and Epidemiology Institute of Preventive Medicine, Academy of Sciences); Chongqing clinical isolates CCS9801, CCS9802, CCS9803, CCS9806, CCS9809, CCS9810, CCS9813, which were identified and preserved by the Department of Clinical Microbiology and Immunology; Mongolia gerbil adapted H pylori strains (abbreviated MG), which were gained from the following steps: gastric mucosal specimens of Mongolia gerbils infected by clinical isolate CCS9803 were inoculated onto nonselective brain heart infusion agar containing 6% horse blood and incubated at 37 °C under microaerobic conditions. After being identified, the positive H pylori strains were named as the first generation of Mongolia gerbil adapted H pylori strains (abbreviation MG1) and then were subcultured with healthy Mongolia gerbil to generate MG2, and in turn to gain the ninth generation (abbreviation MG9). The cultures of all strains mentioned above were preserved at -70 °C in H pylori liquid medium containing 30% glycerol. E.coli DH5α was used as host for cloning DNA and grew in Luria-Bertani (LB) medium at 37 °C. Ampicillin 100 μg/mL was chosen as antibiotics. For blue/white selection of recombinants, 25 μL of 5-bromo-4-chloro-3-indolyl-β-D-galactoside (x-gal) with concentration of 40 mg/mL dissolved with dimethylfo-rmamide were spread on the LB agar plates containing the ampicillin.
Ex-Taq enzyme, restriction endonucleases HhaI, HaeIII, NcoI, XhoI, BamHI, DNA marker and pMD18-T vector were purchased from Takara (Japan). Both plasmid Miniprep and Gel Extraction Kit were bought from Omega (USA).
The H pylori strains’ pure cultures were suspended with 1.0 mL distilled water. The suspension was centrifuged at 10000 g for 1 min. After the removal of the supernatant, the pellets were added with 0.5 mL of distilled water and then boiled for 10 min. The supernatants were transferred to another clean Eppendorf and stored at -20 °C.
Oligonucleotide primers were designed and elevated with Netprimer software and synthesized by Takara. The sense primer (23 mer: 5’-ccatgggcagcccgcatattatt-3’ with NcoI site) and antisense primer (21 mer: 5’-ctcgagtcggtttcttttgcc-3’ with XhoI site) were used to amplify a 710-bp segment of the H pylori adhesin subunit gene hpaA.
Template DNA was added to a 100 μL reaction mixture containing 10 μL 10×Ex-Taq buffer, 0.25 mmol/L MgCl2, 0.025 mmol/L each of the deoxynucleotide phosphates, 2.5 U Ex-Taq polymerase, and 0.25 μmol/L each of the two oligonucleotide primers. The mixtures were overlaid with 30 μL of light mineral oil to prevent evaporation. PCR was performed with an automatic thermal cycler (MiniCyclerTM). The ampli-fication cycle consisted of a pre-denaturation of the target DNA at 94 °C for 3 min, initial denaturation at 94 °C for 50 s, annealing at 60 °C for 50 s, and DNA chain extension at 72 °C for 50 s, with 35 cycles. The final cycle included 10 additional minutes at 72 °C to ensure full extension of product, followed by rapid cooling to 4 °C.
PCR product of 710 bp was recovered and then cloned to pMD18-T vector, then the positive clone was transformed to E.coli DH5α. Extracted plasmid was digested with Nco I and XhoI simultaneously, and the small fragment was recovered to RFLP analysis.
Using NCTC11637 (GenBank accession number X92502) as reference to search endonuclease site within 710-bp hpaA, restriction enzymes Hha I and Hae III were selected. The 8 μL recovered products were subjected to restriction endonuclease either HhaI or HaeIII in a 20 μL reaction volume, with conditions recommended by the manufacturer. Restriction fragments were resolved by 4% agarose gel, then stained with ethidium bromide for about 20 min and washed with distilled water.
The recombinant plasmids pMD18-T-hpaA were sequenced by Takara Corporation with auto-sequencer.
PCR was performed for each of the 12 strains. In all strains, the 710-bp PCR products were successfully amplified. In order to gain pure gene hpaA, 710-bp PCR product was cloned to pMD18-T vector, and then the recombinant plasmids were digested simultaneously with NcoI and XhoI to recover the small fragments.
The objective fragments from 12 different H pylori strains digested with HaeIII could be seen as four types and five types with HhaI. So the hpaA RFLP patterns could separate the 12 H pylori strains into five groups: group I: NCTC11637 and SS1; group II: CCS9809, which RFLP type digested with HaeIII was the same as strains of group I, but HhaI RFLP showed difference compared with the other groups; group III: CCS9810; group IV, CCS9803; group V: CCS9801, CCS9802, CCS9806, CCS9813, MG1, MG3 and MG9 (Figure 1).
Table 1 shows the sequence of gene hpaA of 12 H pylori strains. It could be seen that change of base pair was various in the third site of triad codon but no variation in the conservative KRTIQK, the sixth amino acid of sialyllactose-binding motif.
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | |
| 11637.SEQ | CCATGGGCAG | CCCGCATATT | ATTGAAACCA | ATGAAGTCGC | TTTGAAATTG | AATTACCATC | CAGCTAGCGA |
| CCS9801.SEQ | .......... | .........C | .......... | .......... | ......G... | .......... | T......... |
| CCS9802.SEQ | .......... | .........C | .......... | .......... | ......G... | .......... | T......... |
| CCS9803.SEQ | .......... | .......... | .......... | .......... | ......G... | .......... | .......... |
| CCS9806.SEQ | .......... | .........C | .......... | .......... | ......G... | .......... | T......... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......... | .......... | ........A. | ......G... | .......... | .......... |
| CCS9813.SEQ | .......... | .......... | .......... | .......... | ......G... | .......... | T......... |
| MG1.SEQ | .......... | .......... | .......... | .......... | ......G... | .......... | T......... |
| MG3.SEQ | .......... | .......... | .......... | .......... | ......G... | .......... | T......... |
| MG9.SEQ | .......... | .......... | .......... | .......... | ......G... | .......... | T......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 80 | 90 | 100 | 110 | 120 | 130 | 140 | |
| 11637.SEQ | GAAAGTTCAA | GCGTTAGATG | AAAAGATTTT | GCTTTTAAGG | CCAGCTTTCC | AATATAGCGA | TAATATCGCT |
| CCS9801.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| CCS9802.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| CCS9803.SEQ | .......... | .......... | .......... | A.......A. | ..A.....T. | ....C..... | ......T... |
| CCS9806.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......... | .......... | A.......A. | ..A.....T. | ....C..... | ......T... |
| CCS9813.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| MG1.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| MG3.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| MG9.SEQ | .......... | .......... | ........C. | A......... | ..G.....T. | ....C..... | ......T... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 150 | 160 | 170 | 180 | 190 | 200 | 210 | |
| 11637.SEQ | AAAGAGTATG | AAAACAAATT | CAAGAATCAA | ACCGCGCTCA | AGGTTGAACA | GATTTTGCAA | AATCAAGGCT |
| CCS9801.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| CCS9802.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| CCS9803.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| CCS9806.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .....G.... |
| CCS9813.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| MG1.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| MG3.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| MG9.SEQ | .......... | .......... | .......... | .....A.... | .......... | .......... | .....G.... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 220 | 230 | 240 | 250 | 260 | 270 | 280 | |
| 11637.SEQ | ATAAGGTTAT | TAGCGTAGAT | AGCAGCGATA | AAGACGATTT | TTCTTTTGCA | CAAAAAAAAG | AAGGGTATTT |
| CCS9801.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| CCS9802.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| CCS9803.SEQ | .......... | ..AT...... | ..T....... | ........C. | .........G | .......... | .......... |
| CCS9806.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| CCS9809.SEQ | .......... | .......... | ........... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| CCS9813.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| MG1.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| MG3.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| MG9.SEQ | .......... | ..AT...... | .......... | ........C. | .........G | .......... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 290 | 300 | 310 | 320 | 330 | 340 | 350 | |
| 11637.SEQ | GGCGGTTGCT | ATGAATGGCG | AAATTGTTTT | ACGCCCCGAT | CCTAAAAGGA | CCATACAGAA | AAAATCAGAA |
| CCS9801.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| CCS9802.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| CCS9803.SEQ | ...C..C... | ....G..... | .......... | G......... | .......... | .......... | .......... |
| CCS9806.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| CCS9809.SEQ | .......... | .......... | .........C | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | |
| CCS9813.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| MG1.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| MG3.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| MG9.SEQ | ...C..C... | ....G..... | .......... | .......... | .......... | .......... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 360 | 370 | 380 | 390 | 400 | 410 | 420 | |
| 11637.SEQ | CCCGGGTTAT | TATTCTCCAC | CGGTTTGGAC | AAAATGGAAG | GGGTTTTAAT | CCCGGCTGGG | TTTATTAAGG |
| CCS9801.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| CCS9802.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| CCS9803.SEQ | .......... | .......... | T........T | .......... | .......... | .......... | .....C.... |
| CCS9806.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| CCS9813.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| MG1.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| MG3.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| MG9.SEQ | .......... | .......... | T........T | .......... | .......... | ......C... | .....C.... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 430 | 440 | 450 | 460 | 470 | 480 | 490 | |
| 11637.SEQ | TTACCATACT | AGAGCCTATG | AGTGGGGAAT | CTTTGGATTC | TTTTACGATG | GATTTGAGCG | AGTTGGACAT |
| CCS9801.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| CCS9802.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| CCS9803.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .......... | .......... |
| CCS9806.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | ........T. | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9813.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| MG1.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| MG3.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| MG9.SEQ | ........T. | .......... | .......... | ....A..... | .......... | .....A.... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 500 | 510 | 520 | 530 | 540 | 550 | 560 | |
| 11637.SEQ | TCAAGAAAAA | TTCTTAAAAA | CCACCCATTC | AAGCCATAGC | GGGGGGTTAG | TTAGCACTAT | GGTTAAGGGA |
| CCS9801.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9802.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9803.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9806.SEQ | C......... | .......... | .......... | .......... | .......... | ........G. | .......... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9813.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| MG1.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| MG3.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| MG9.SEQ | C......... | .......... | .......... | .......... | .......... | .......... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 570 | 580 | 590 | 600 | 610 | 620 | 630 | |
| 11637.SEQ | ACGGATAATT | CTAATGACGC | GATCAAGAGC | GCTTTGAATA | AGATTTTTGC | AAATATCATG | CAAGAAATAG |
| CCS9801.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| CCS9802.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| CCS9803.SEQ | .......... | .......T.. | .......... | ........C. | .......... | .......... | .......... |
| CCS9806.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......T.. | .........T | .......... | .......... | .......... | .......... |
| CCS9813.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| MG1.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| MG3.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| MG9.SEQ | .......... | .......T.. | .......... | .......... | .......... | ..G....... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 640 | 650 | 660 | 670 | 680 | 690 | 700 | |
| 11637.SEQ | ACAAAAAACT | CACTCAAAAG | AATTTAGAAT | CTTATCAAAA | AGACGCCAAA | GAATTAAAAG | GCAAAAGAAA |
| CCS9801.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| CCS9802.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| CCS9803.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9806.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| CCS9809.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| CCS9810.SEQ | .......... | .......... | .......... | .......... | .......... | .....G.... | .......... |
| CCS9813.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| MG1.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| MG3.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| MG9.SEQ | .......... | .......... | .......... | .......... | ......T..G | .......... | .......... |
| SS1.SEQ | .......... | .......... | .......... | .......... | .......... | .......... | .......... |
| 710 | |||||||
| 11637.SEQ | CCGACTCGAG | ||||||
| CCS9801.SEQ | .......... | ||||||
| CCS9802.SEQ | .......... | ||||||
| CCS9803.SEQ | .......... | ||||||
| CCS9806.SEQ | .......... | ||||||
| CCS9809.SEQ | .......... | ||||||
| CCS9810.SEQ | .......... | ||||||
| CCS9813.SEQ | .......... | ||||||
| MG1.SEQ | .......... | ||||||
| MG3.SEQ | .......... | ||||||
| MG9.SEQ | .......... | ||||||
| SS1.SEQ | .......... |
To assess the variation of 12 gene hpaA of different H pylori strains, the sequence data were analyzed by DNAsis software. From the phylogenetic tree (Figure 2), it could be seen that: (1) The homologies of base pair and amino acid sequence in strains NCTC11637, SS1, CCS9809 were 99.6% and 98.9%, respectively; (2) The homologies of base pair and amino acid sequence between CCS9803 and CCS9810 were 97.7% and 99.1%; (3) That of the rest strains, CCS9801, CCS9802, CCS9806, CCS9813, MG1, MG3, MG9 could hit 99.4% and 98.4%; (4) The base pair homologies in all hpaA fragments of different sources were more than 94.6%, therefore the correspondence of deduced amino acid sequence was higher than 96.8% between each other.
Special adhesion of bacteria to host cells was conducted by adhesins and receptors system, which relied on combination of some amino acids with their special receptors. The adhesion and colonization was one of the most basic factors of H pylori infection. The colonization elements included dynamic, urease enzyme and adhesins, etc.[7]. The reason that H pylori could not be eliminated with foods via stomach’s peristalsis was related to its close adherence to stomach epithelial cells[8,9]. Histological studies revealed that H pylori could be found only in stomach epithelial cells, and generally existed at antrum of stomach. H pylori had strong adherence to stomach epithelial cells, but also with host, tissue and obvious sites specificity. Different H pylori strains revealed transparent discrepancy while adhering to the same gastric mucosa. All these indicated a complex system of adhesins and receptors existing between host and H pylori[6,10,11]. Evan et al[12], found that neuraminyl lactose binding hemagglutinin (NLBH or hpaA) of H pylori could closely bind to special receptors on human stomach mucosa. The special receptors, which included MG3 ganglioside, sulfate lactosylceramide, could help H pylori tightly adhere to stomach epithelial cells and further injure stomach mucosa.
Evans et al[13], using restriction endonucleases Sau3A and Hinf I to digest the gene hpaA, a 375-bp polymerase chain reaction-amplified product, detected that seven different polymorphic types were found in hpaA, which was obtained from 50 different H pylori isolates. By gene sequencing, more than 90% base substitutions were very conservative, they either did not change the encoded amino acid or substituted a homologous amino acid, and caused no detectable antigenic or functional effect on hpaA.
In this study a 710-bp polymerase chain reaction amplified sequence of hpaA, obtained from 12 different H pylori strains was digested with HhaI and HaeIII respectively and analyzed with agarose gel electrophoresis. The result revealed that there were four RFLP polymorphic types with HaeIII and five with HhaI, so we could divide 12 H pylori strains into five groups, which were (1) NCTC11637, SS1; (2) CCS9809; (3) CCS9810; (4) CCS9803; (5) CCS9801, CCS9802, CCS9806, CCS9813, MG1, MG3, MG9. There was no difference in RFLP between NCTC11637 and SS1, and both were accordant to the predictive data of GenBank. On the other hand, hpaA RFLP analysis of Chongqing clinical H pylori isolates CCS9810, CCS9813 and CCS9803 were different between each other and distinguished from standard strains. Result of CCS9809 digested with HaeIII was the same as with NCTC11637 and SS1, but different from other strains by HhaI digestion. It indicated that, firstly, there existed remarkable difference in hpaA RFLP among Chinese and international standard strains. Secondly, it showed difference in HhaI and HaeIII recognizing sites of gene hpaA from Chinese clinical isolates, which were obtained from different gastrointestinal diseases. These proved that there was numerous polymorphism of gene hpaA at nucleic acid level.
While setting up infected animal model, in order to help H pylori colonizing at stomach mucosa, we pretreated Mongolia gerbil with medicine to injure its stomach mucosa and then subcultured live H pylori. Strains MG1 to MG8 were gained in turn with this method but strain MG9 was obtained through directly subculturing live H pylori without pretreating Mongolia gerbil stomach mucosa with medicine. This indicated that the colonizing ability of H pylori had been enhanced in the progress of continuous infection. Besides, the types of hpaA RFLP of three MG strains were the same between one another, but different from CCS9803. Therefore, it might be variation of H pylori during adaptation to a different host. Since the RFLP type of CCS9801, CCS9802, CCS9806 and CCS9813 was the same as MG strains, it might suggest that these clinical isolates originated from animals.
In conclusion, RFLP analysis of gene hpaA digested with HhaI and HaeIII was a good means to differentiate H pylori strains and to study transmission route of H pylori at molecular level. Furthermore we could utilize the means to study H pylori’s distribution, epidemic characteristics and molecular epidemiology.
We thank Qing-Hua Xie for culturing H pylori strains. We thank Dr. Guo-Ping Li for helpful discussions and for writing the manuscript.
| 1. | Ye W, Held M, Lagergren J, Engstrand L, Blot WJ, McLaughlin JK, Nyrén O. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J Natl Cancer Inst. 2004;96:388-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 256] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 2. | Toyokawa T, Yokota K, Mizuno M, Fujinami Y, Takenaka R, Okada H, Hayashi S, Hirai Y, Oguma K, Shiratori Y. Characterization of elongated Helicobacter pylori isolated from a patient with gastric-mucosa-associated lymphoid-tissue lymphoma. J Med Microbiol. 2004;53:207-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 3. | Lundström AM, Blom K, Sundaeus V, Bölin I. HpaA shows variable surface localization but the gene expression is similar in different Helicobacter pylori strains. Microb Pathog. 2001;31:243-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Nasrolahei M, Maleki I, Emadian O. Helicobacter pylori colonization in dental plaque and gastric infection. Rom J Gastroenterol. 2003;12:293-296. [PubMed] |
| 5. | Lu XL, Qian KD, Tang XQ, Zhu YL. Distribution of H.pylori antigens in gastric mucosa and its significance. J Zhejiang Univ Sci. 2004;5:242-245. [PubMed] [DOI] [Full Text] |
| 6. | Blom K, Lundin BS, Bölin I, Svennerholm A. Flow cytometric analysis of the localization of Helicobacter pylori antigens during different growth phases. FEMS Immunol Med Microbiol. 2001;30:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Müller I, Medina-Selby A, Palacios JL, Martinez P, Opazo P, Bruce E, Mancilla M, Valenzuela P, Yudelevich A, Venegas A. Cloning and comparison of ten gene sequences of a Chilean H. pylori strain with other H. pylori strains revealed higher variability for VacA and CagA virulence factors. Biol Res. 2002;35:67-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Papadaki HA, Skordilis P, Minadakis G, Roussomoustakaki M, Katrinakis G, Psyllaki M, Tzardi M, Kouroumalis E, Eliopoulos GD. Complete regression of primary gastric plasmacytoma following Helicobacter pylori eradication. Ann Hematol. 2003;82:589-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Wabinga HR. Comparison of immunohistochemical and modified Giemsa stains for demonstration of Helicobacter pylori infection in an African population. Afr Health Sci. 2002;2:52-55. [PubMed] |
| 10. | Voland P, Hafsi N, Zeitner M, Laforsch S, Wagner H, Prinz C. Antigenic properties of HpaA and Omp18, two outer membrane proteins of Helicobacter pylori. Infect Immun. 2003;71:3837-3843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Lundstrom A, Bolin I, Bystrom M, Nystrom S. Recombinant HpaA purified from Escherichia coli has biological properties similar to those of native Helicobacter pylori HpaA. APMIS. 2003;111:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Evans DG, Evans DJ, Moulds JJ, Graham DY. N-acetylneuraminyllactose-binding fibrillar hemagglutinin of Campylobacter pylori: a putative colonization factor antigen. Infect Immun. 1988;56:2896-2906. [PubMed] |
| 13. | Evans DG, Evans DJ, Lampert HC, Graham DY. Restriction fragment length polymorphism in the adhesin gene hpaA of Helicobacter pylori. Am J Gastroenterol. 1995;90:1282-1288. [PubMed] |