Published online May 7, 2005. doi: 10.3748/wjg.v11.i17.2609
Revised: October 8, 2004
Accepted: November 19, 2004
Published online: May 7, 2005
AIM: To explore the possibility of the replacement of the gag gene between human immunodeficiency virus and bovine immunodeficiency virus, to achieve chimeric virions, and thereby gain a new kind of AIDS vaccine based on BHIV chimeric viruses.
METHODS: A series of chimeric BHIV proviral DNAs differing in the replacement regions in gag gene were constructed, and then were transfected into 293T cells. The expression of chimeric viral genes was detected at the RNA and protein level. The supernatant of 293T cell was ultra centrifuged to detect the probable chimeric virion. Once the chimeric virion was detected, its biological activities were also assayed by infecting HIV-sensitive MT4 cells.
RESULTS: Four chimeric BHIV proviral DNAs were constructed. Genes in chimeric viruses expressed correctly in transfected 293T cells. All four constructs assembled chimeric virions with different degrees of efficiency. These virions had complete structures common to retroviruses and packaged genomic RNAs, but the cleavages of the precursor Gag proteins were abnormal to some extent. Three of these virions tested could attach and enter into MT4 cells, and one of them could complete the course of reverse transcription. Yet none of them could replicate in MT4 cells.
CONCLUSION: The replacement of partial gag gene of HIV with BIV gag gene is feasible. Genes in chimeric BHIVs are accurately expressed, and virions are assembled. These chimeric BHIVs (proviral DNA together with virus particles) have the potential to become a new kind of HIV/AIDS vaccine.
- Citation: Zhu YX, Liu C, Liu XL, Qiao WT, Chen QM, Zeng Y, Geng YQ. Construction and characterization of chimeric BHIV (BIV/HIV-1) viruses carrying the bovine immunodeficiency virus gag gene. World J Gastroenterol 2005; 11(17): 2609-2615
- URL: https://www.wjgnet.com/1007-9327/full/v11/i17/2609.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i17.2609
The HIV epidemic continues to expand at an alarming rate and is predicted to be the worst infectious disease ever to affect human beings, and the situation in Asia is also alarming[1]. Historical experience indicates that vaccines continue to be the most cost efficient and effective intervention available for preventing infectious diseases.
Up to now, there have been 80 candidate HIV/AIDS vaccines. Forty-seven of them have completed trials, but found to be either unsafe or ineffective, while 33 of them are in ongoing trials: one in phase III, three in phase II, two in phase I/II, and the rest in phase I. These vaccines include virus-like particles, peptides, recombinant protein subunits, recombinant bacterial vectors, recombinant viral vectors, and DNA vaccines[2]. However, the chances of getting an effective HIV/AIDS vaccine with current approaches are still very negligible. We still need new vaccine strategies against HIV/AIDS[3].
The chimeric HIV might just be such a new kind of HIV/AIDS vaccine, because it can mimic the natural infection of HIV[4-6], but out of concern about safety, current chimeric SHIVs are unfit for vaccines[5-7]. Actually, they are used as challenge viruses in AIDS/rhesus models to evaluate the efficacy of HIV/AIDS vaccines. HIV-1 and SIV are so closely related phylogenetically that the chimeric virus of SIV/HIV is too dangerous to be a vaccine[5,6]. Then how about a chimeric virus between HIV and non-primates lentivirus, such as the bovine immunodeficiency virus?
Bovine immunodeficiency virus (BIV) is a lentivirus, which resembles HIV in its structural, genomic, antigenic, and biological properties[8]. Unlike HIV, BIV is a relatively mild lentivirus, and never causes severe acquired immune deficiency syndrome in its host, the cow[9,10]. BIV seropositivity has been correlated with decreased milk production in dairy cattle, but has not been directly linked with clinical disease in naturally infected cattle[11]. Considering the similarity of genome to HIV and its low pathogenicity, BIV may be an appropriate candidate to combine with HIV to create a new kind of HIV chimeric virus - BHIV.
Primary work showed the expression of BIV gag-pol gene in HIV backbone in human original MT4 cells[12]. This time we focus on the gag gene. We tried to explore the possibility of the replacement of the gag gene between human immunodeficiency virus and BIV. In doing so, a series of chimeric BHIV (BIV/HIV-1) proviruses carrying the BIV gag gene have been constructed. In order to improve the expression of chimeric genes in human cells, the CMV promoter is introduced.
Infectious cDNA of BIV127 and HIV-1 HXBc2 were kept in our laboratory, and pcDNA3.1(-) vector was from Invitrogen.
Construction of chimeric BHIV proviral DNA Modification of pcDNA3.1(-) vector: The pcDNA3.1(-) plasmid was double digested with EcoRI and BamHI, then the ends were blunted with T4 DNA polymerase and self-ligated by T4 DNA ligase, so that only one SacI site remained. Then the BssHII site was removed by cutting, blunting and religating.
Replacement of 5’ HIV LTR U3 region by CMV promoter: The fragment 1 of 5’ HIV LTR R U5 and partial gag was amplified by PCR, using sense primer from HIV-1 HXBc2 positions 441-459 nt (5’ TCGAC-TTTTGCCTGTACTGGGTCT 3’), and anti-sense primer from HIV-1 HXBc2 positions 2 045-2 026nt (5’ TCCTTTCCACATTTCCAACA3’). The fragment 2 of 3’ HIV LTR and partial env was amplified by PCR, using sense primer from HIV-1 HXBc2 positions 8824-8842nt (5’ GTGATTGGATGGCCTACTG 3’) and anti-sense primer from HIV-1 HXBc2 positions 9 719-9700nt (5’ TGCTAGAGATTTTCCACACT 3’).
Modified pcDNA3.1(-) vector was excised with SacI, blunted, excised with ApaI, then was ligated with the PCR product of fragment 1, which was already digested with ApaI. Thus, the 5’ HIV LTR U3 region was replaced by the CMV promoter of the modified pcDNA3.1(-) vector. We got the first intermediate plasmid pCMV1.
Plasmid pCMV1 was double excised with XhoI and EcoRV, which was ligated with the PCR product of fragment 2, which was already digested with XhoI. Thus, the 3’ HIV LTR was transferred into the vector. We got the second intermediate plasmid pCMV2.
Plasmid pCMV2 was double digested with ApaI and XhoI, and was ligated with the 7-kb HIV ApaI-XhoI fragment, then the complete genome of HIV was transferred to the modified pcDNA3.1(-) vector, along with the transfer of CMV promoter to the 5’ HIV LTR U3 region. It was named pCHIV.
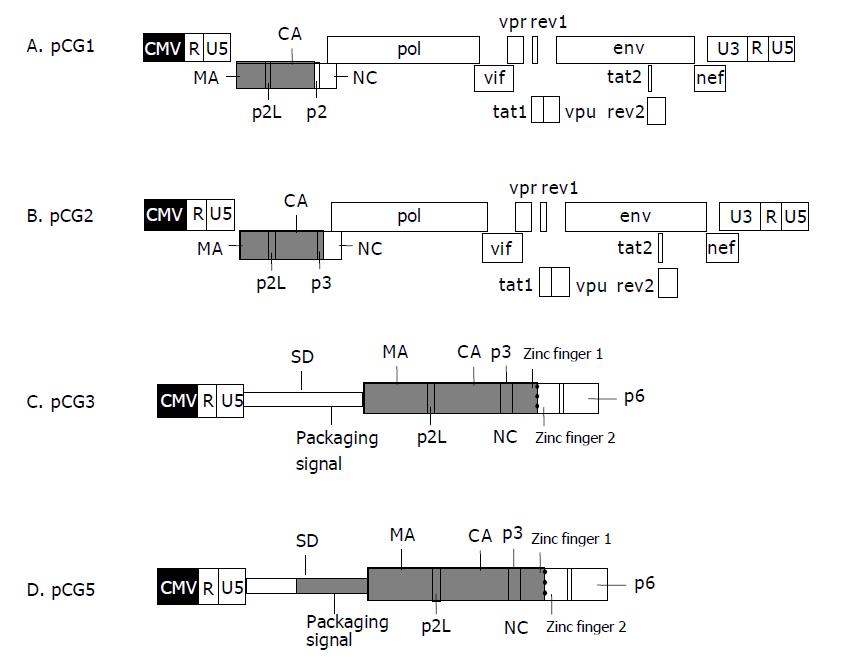
Construction of pCG1 and pCG2: Chimeric gag genes of pCG1 and pCG2 were derived from C2 and C3[13]. The chimeric gag genes from C2 and C3 were excised with BssHII/ApaI, and ligated into the vector of pCHIV, which was already digested with BssHII/ApaI. So we got the complete genome of chimeric HIV in pCG1 and pCG2.
Construction of pCG3: Two rounds of PCR produced the chimeric BHIV gag fragments of pCG3. The first round PCR used BIV127 as template, and the product of the first round together with pCG1 were used as templates in the second round PCR.
First round PCR sense primer: 5’ TAAGGTTAGGGTGACAC 3’, from BIV127 positions 741-757nt. Anti-sense primer: 5’ CC-TACAATTCCTCTTCAAATG 3’, from BIC127 positions 1 963-1945nt.
Second round PCR sense primer: 5’ TCGAC-TTTTGCCTGTACTGGGTCT 3’, from HIV-1 HXBc2 positions 441-459nt. Anti-sense primer was same as in the first round PCR.
The complete genome of pCG3 proviral DNA was constructed as follows: pCHIV was excised with ApaI, blunted, excised with BssHII, then ligated with the product of the second round PCR that was already digested with BssHII.
Construction of pCG5: The proviral genome DNA was constructed by blunt end ligation. Plasmid pCHIV was double-digested with BssHII/ApaI. The ends were blunted with T4 DNA polymerase, and then were dephosphorylated by calf intestine alkaline phosphatase. The inserted fragment was a PCR product that was already phosphorylated by T4 polynucleotide kinase. After ligation, the clone of corrected insert orientation was selected by restriction enzyme mapping.
The sense primer of the PCR was 5’ CAGAAGACTCCGGACAG 3’, from BIV127667-683nt. Anti-sense primer was 5’ CC-TACAATTCCTCTTCAAATG 3’, from BIC127 1963-1945nt.
In a level 3 biosafety (P3) laboratory, four BHIV clones were individually transfected into 40-50% confluence 293T cells that were grown in 10% FBS DMEM in a six-well plate by using Polyfect (Qiagen). In order to reduce the side effect of remnant proviral DNA and Polyfect, cells were washed with PBS thrice and fresh culture medium was added 24 h after transfection. The cell RNA and protein were extracted 72 h post-transfection by Tri reagent (Sigma).
To perform electron microscopy (EM), supernatants were fixed with 2.5% glutaraldehyde, followed by treatment with 4% osmium tetroxide, and then routinely processed. Samples were stained with lead citrate and uranyl acetate and then visualized by using a JEOL JEM-2000 FX transmission electron microscope.
The culture supernatant was collected 72 h post-transfection and filtered with 0.45-μm filter to remove cell fractions. The cell-free supernatant was ultra centrifuged at 30000 g for 3 h. The pellet was stored at -80 °C.
In Western blot, anti-BIV capsid (CA) polyclone antibody was used to detect the chimeric Gag precursor and mature CA protein in these virions. Reverse transcriptase activities in these virions were assayed by Reverse Transcriptase Assay colorimetric kit (Roche). The genomic RNA in these virions were also extracted as follows: virion pellets were re-suspended, DNase added to digest the remnant proviral DNA at 37 °C for 30 min, then the mixture was extracted with phenol/chloroform and RNA precipitated with ethanol.
The culture supernatant was collected 72 h post-transfection and filtered with 0.45-μm filter to remove cell fractions. The cell-free supernatant was added into 5×106 in 5 mL 10% FBS RPMI-1640 medium. After incubation at 37 °C for 18 h, MT4 cells were washed thrice with PBS, then fresh medium was added and returned to incubator. After 8 h, cells were again washed thrice with PBS, and then their RNA, DNA and protein were extracted by Tri reagent (Sigma).
RT-PCR was performed to detect the early (tat gene) and late (gag gene) RNA transcripts in transfected 293T cells. RT-PCR was also performed to detect the genomic RNA (env gene) as a representative (Table 1).
| Primer name | Sequence | Location | Product length |
| tat RT-PCR up | 5’-GACTCGGCTTGCTGAAG-3’ | HIV 695-711 | 250 bp |
| tat RT-PCR low | 5’-GCTGTCTCCGCTTCTTC-3’ | HIV 5993-5977 | |
| gag RT-PCR up | 5’-GGAGGCCAGAGCTGATAAG-3’ | BIV 1044-1062 | 560 bp |
| gag RT-PCR low | 5’-GTCTGTGTACGGCTCCTTG-3’ | BIV1608-1590 | |
| env PCR up | 5’-AATGACGCTGACGGTACAGG-3’ | HIV 7826-7845 | 660 bp |
| env PCR low | 5’-GTGCCAAGGATCCGTTCAC-3’ | HIV 8487-8469 |
Four chimeric BHIV proviral DNAs were constructed. Both restriction enzyme mapping and DNA sequencing confirmed the sequences of these proviruses. The genomic structure of each provirus is illustrated in Figure 1.
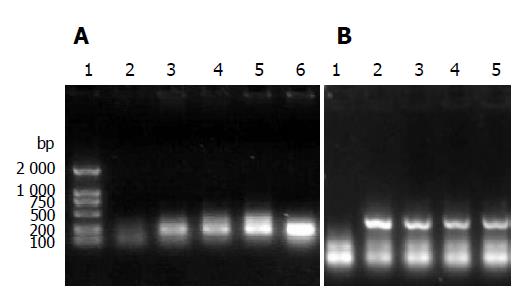
After transfection, RNA from 293T cells was extracted. The detection of early transcript of tat gene and late transcript of gag gene was performed by RT-PCR. Both early and late gene transcripts of RNA were detected in all four chimeric viruses. The results of RT-PCR are shown in Figure 2.
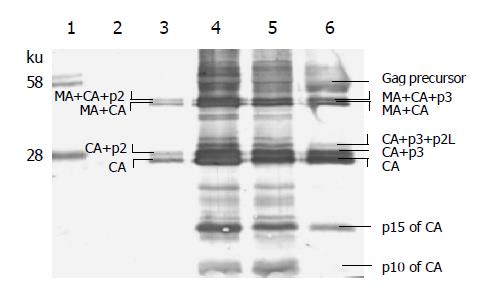
Western blot analysis (Figure 3) showed that in 293T cells all four chimeric Gag precursors were expressed. There was no significant difference in the amount of Gag expression. All fours Gag precursors were partially cleaved. There were two cleavage profiles among these four Gag precursors: pCG1 belonged to one cleavage profile; pCG2, pCG3, pCG5 belonged to another. The spectrum of the latter profile was depicted on the right side of Figure 3.
The cell-free supernatant was visualized by electron microscopy. Figure 4 shows the results observed by EM. All four chimeric proviruses assembled virions in 293T cells. These virions have a diameter of about 100 nm, the typical scale of lentivirus. Unlike mature HIV virions, these particles do not have spindle cores, indicating that they are in the immature status.
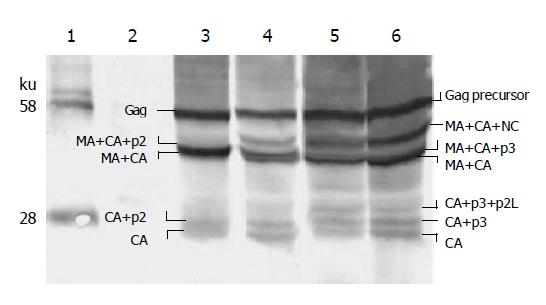
The cell-free supernatant was ultra centrifuged. The pellet of viral particles was analyzed by Western blot analysis, as shown in Figure 5. As it shows, very few Gag precursors in virions were left, but the cleavage was still not complete. A large proportion of BIV CA existed in intermediate cleavage products, especially in CA+p3. A small part of BIV CA was still cut by HIV-1 protease and the products of p15 and p10 of CA were generated.
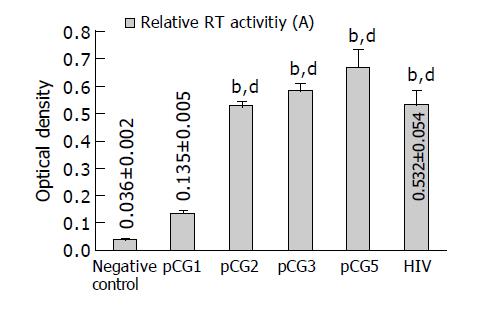
The relative RT activities of four chimeric virions are shown in Figure 6. Each measure was taken with virions centrifuged from 1 mL supernatant of 293T cells. The transfection efficiency and cell confluence will also affect the release of chimeric virions, and the data of virion RT activities were semi-quantitative. We can conclude that the RT activity of pCG1 virion was lower than that of HIV positive control (P<0.01 vs HIV), the other three do not have any significant difference from that of HIV control.
Although the package signals and the package signal recognizers (mainly the nucleic capsid, NC, in Gag) were different in these four chimeric virions, they all included genomic RNAs as the results of RT-PCR shown in Figure 7.
Virions of pCG2, pCG3 and pCG5 in supernatant were incubated with human lymphocyte primary MT4 cells for 24 h, then the remnant virions and culture medium were carefully eliminated. The protein of chimeric Gag from virions can be detected in MT4 cells using the Western blot analysis shown in Figure 8.
After incubation of chimeric virions with MT4 cells, we carefully eliminated the remnant virions and traced proviral DNA in the culture medium, then extracted the genomic DNA of MT4 cells. Detection of genomic DNA of chimeric virions was performed by PCR. Only in pCG3 virion-infected MT4 cells we can find positive result as illustrated in Figure 9. That means virions of pCG3 can complete reverse transcription in MT4 cells.
The constitutionally strong promoter of CMV can replace the HIV-1 LTR U3 region and promote the RNA transcription of HIV-1. Generally speaking, the transcription efficiency of the CMV promoter is higher than that of HIV-1 LTR in human cells[14]. So, the replacement of promoter will enhance the expression of chimeric viral genes. Incidentally, the replacement took place only on the 5’ LTR U3 region, not on the 3’ LTR U3 region, nor any of LTR. That means the replacement only promotes the quantity of transcription, and does not change the quality of transcripts.
The expression of early genes in HIV relies on the correct post-transcriptional splicing of RNA in the nucleus of host cells[15]. In the genome of pCG4, the splicing donor comes from BIV, while the splicing receptor belongs to HIV-1. From the result of tat gene RT-PCR (Figure 2A), we can see that the heterogeneous donor and receptor matched each other and generated correct splicing.
The chimeric gag gene as a late gene can be transcribed and translated in 293T cells. That means the early gene can work and trigger the expression of late genes[16]. The expression of Gag precursor in four chimeric BHIVs was of the same magnitude, and all four chimeric BHIVs assembled virions. However, the virions assembled in pCG1 were much less than the other three. Some research pointed out that the p2 plays an important role in HIV-1 virion assembly[17-19]. The results here suggest that p3 - the counterpart of p2 - in BIV Gag is also important in efficient assembly of BIV Gag. Virions of chimeric Gag (BIV Gag as a major part) can still be assembled if BIV p3 is replaced by HIV-1 p2, but the assembly efficiency will be very low. This is not congruent with the result of Guo et al[13]. We deduce that this is because of the high transcription efficiency of CMV promoter counteracting the low assembly efficiency of pCG1 Gag.
The primate lentivirus proteases have been intensively investigated[20], and it was found that the specificity is conserved among HIV-1, HIV-2, and SIV. For instance, synthetic HIV-2 protease cleaves the Gag precursor of HIV-1 with the same specificity as HIV-1 protease[21,22], and the proteolytic processing of the HIV-2 Gag precursor is very similar to the processing of the SIVMne Gag precursor[23]. However, the substrate specificity of protease between HIV-1 and the non-primate lentivirus BIV has not been studied previously. In these experiments, the chimeric Gag precursors not only in cells but also in virions were cleaved by HIV-1 protease. Among these cleaving sites in chimeric Gags, some have come from HIV-1 Gag, some from BIV Gag, and some are even mosaics from both HIV-1 and BIV. Our results indicate that all these cleavage sites can be recognized and cleaved by HIV-1 protease although in different efficiency (Figures 3 and 5). The relatively low cleaving efficiency may lead to the immature morphology of virions (Figure 4).
Previous study indicated that a small percentage of the CA protein undergoes secondary proteolysis to generate additional peptides during the natural cleavage of BIV Gag precursor. A 10-kDa C-terminal peptide of CA, has been immunologically identified; another putative 16-ku N-terminal peptide of CA has not been immunologically identified yet[24,25]. From Figure 4 we can see that HIV-1 protease also can recognize and cleave this secondary alternate site inside BIV CA. Besides the alternate CA cleavage product of p10, the other putative product of a 16-ku peptide is also immunologically identified as p15. This adds to the conservative character of protease substrate specificity among lentivirus.
There are four cleavage sites within the HIV-1 Gag precursor (six within chimeric Gag, including the secondary alternate cleavage site inside CA), which on cleavage gives rise to the mature core proteins MA, CA, NA, and p6. (For simplicity and in the absence of specific data in our experiments, we exclude the HIV-1 Gag spacer peptide of p1 between NC and p6 from the discussion.) The cleaving efficiency among the four sites is different. The site of p2-NC was cleaved first, NC-p6 second, MA-CA third, and CA-p2 last[26]. Sequential cleavage of Gag precursor generates certain kinds of intermediates, and from the profile of intermediates, we can deduce the cleavage sequence. In the precursor of pCG1 Gag, junctions of p2-NC and NC-p6 have come from HIV-1; they can surely be cleaved by HIV-1 protease efficiently. So they are the first and second cleavage sites. The junction of CA-p2 is a chimeric junction; the segment to the left of scissible bond comes from BIV, to the right of the scissible bond comes from HIV-1. It is the last site cleaved, and is responsible for the accumulation of CA+p2 intermediate (Figure 5). In the precursor of pCG2 or pCG3 or pCG5, the p3-NC junction no longer comes from HIV-1. So, the cleavage efficiency is reduced, and a new intermediate of MA+CA +NC emerged (Figure 3). The deduced cleavage sequence of chimeric Gag precursor is illustrated in Figure 10.
The structure of these chimeric virions is complete. They have genomic RNA, reverse transcriptase, core proteins, and envelope together with glycoprotein of gp120 and gp41 (this is deduced from the fact that they can attach and enter into MT4 cells). One of them even can complete reverse transcription in MT4 cells. But none of them can replicate in MT4 cells. Because of these characteristics, they have the potential of being a new kind of HIV AIDS vaccine. They are relatively safe. They can express HIV antigen efficiently. They can assemble virions, and these virions can enter human lymphocytes. We anticipate that they can mimic the natural infection of HIV and stimulate the human immune system to elicit both cellular and humoral immune responses against HIVs.
In summary, the replacement of partial gag gene of HIV with BIV gag gene is feasible. Genes in chimeric BHIVs are accurately expressed, and virions are assembled. Chimeric BHIVs (proviral DNA together with virus particles) are expected to be a new kind of HIV/AIDS vaccine candidate.
We thank Liang Chen for providing the plasmid DNA of C2 and C3.
Co-correspondents: Yi Zeng
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Roger D. HIV Surveillance, Prevention, Intervention, and treatment in Asia. The XV International HIV/AIDS Conference. New York: Guilford Press 2004; . |
| 2. | Available from: http: //www.iavireport.org/trialsdb/default.asp. |
| 3. | Berzofsky JA, Ahlers JD, Janik J, Morris J, Oh S, Terabe M, Belyakov IM. Progress on new vaccine strategies against chronic viral infections. J Clin Invest. 2004;114:450-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Ui M, Kuwata T, Igarashi T, Ibuki K, Miyazaki Y, Kozyrev IL, Enose Y, Shimada T, Uesaka H, Yamamoto H. Protection of macaques against a SHIV with a homologous HIV-1 Env and a pathogenic SHIV-89.6P with a heterologous Env by vaccination with multiple gene-deleted SHIVs. Virology. 1999;265:252-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Silverstein PS, Mackay GA, Mukherjee S, Li Z, Piatak M, Lifson JD, Narayan O, Kumar A. Pathogenic simian/human immunodeficiency virus SHIV(KU) inoculated into immunized macaques caused infection, but virus burdens progressively declined with time. J Virol. 2000;74:10489-10497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | Kumar A, Lifson JD, Li Z, Jia F, Mukherjee S, Adany I, Liu Z, Piatak M, Sheffer D, McClure HM. Sequential immunization of macaques with two differentially attenuated vaccines induced long-term virus-specific immune responses and conferred protection against AIDS caused by heterologous simian human immunodeficiency Virus (SHIV(89.6)P). Virology. 2001;279:241-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Whitney JB, Ruprecht RM. Live attenuated HIV vaccines: pitfalls and prospects. Curr Opin Infect Dis. 2004;17:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Gonda MA. Bovine immunodeficiency virus. AIDS. 1992;6:759-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Gonda MA, Luther DG, Fong SE, Tobin GJ. Bovine immunodeficiency virus: molecular biology and virus-host interactions. Virus Res. 1994;32:155-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Carpenter S, Vaughn EM, Yang J, Baccam P, Roth JA, Wannemuehler Y. Antigenic and genetic stability of bovine immunodeficiency virus during long-term persistence in cattle experimentally infected with the BIV(R29) isolate. J Gen Virol. 2000;81:1463-1472. [PubMed] |
| 11. | McNab WB, Jacobs RM, Smith HE. A serological survey for bovine immunodeficiency-like virus in Ontario dairy cattle and associations between test results, production records and management practices. Can J Vet Res. 1994;58:36-41. [PubMed] |
| 12. | Chen G, Wang S, Xiong K, Wang J, Ye T, Dong W, Wang Q, Chen Q, Geng Y, Wood C. Construction and characterization of a chimeric virus (BIV/HIV-1) carrying the bovine immunodeficiency virus gag-pol gene. AIDS. 2002;16:123-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 13. | Guo X, Hu J, Whitney JB, Russell RS, Liang C. Important role for the CA-NC spacer region in the assembly of bovine immunodeficiency virus Gag protein. J Virol. 2004;78:551-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Paya CV, Virelizier JL, Michelson S. Modulation of T-cell activation through protein kinase C- or A-dependent signalling pathways synergistically increases human immunodeficiency virus long terminal repeat induction by cytomegalovirus immediate-early proteins. J Virol. 1991;65:5477-5484. [PubMed] |
| 15. | Feinberg MB, Jarrett RF, Aldovini A, Gallo RC, Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986;46:807-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 562] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 16. | Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 961] [Cited by in RCA: 1008] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 17. | Kräusslich HG, Fäcke M, Heuser AM, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407-3419. [PubMed] |
| 18. | Accola MA, Höglund S, Göttlinger HG. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072-2078. [PubMed] |
| 19. | Morikawa Y, Hockley DJ, Nermut MV, Jones IM. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J Virol. 2000;74:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 68] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol. 2002;76:10226-10233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Wu JC, Carr SF, Jarnagin K, Kirsher S, Barnett J, Chow J, Chan HW, Chen MS, Medzihradszky D, Yamashiro D. Synthetic HIV-2 protease cleaves the GAG precursor of HIV-1 with the same specificity as HIV-1 protease. Arch Biochem Biophys. 1990;277:306-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Pichuantes S, Babé LM, Barr PJ, DeCamp DL, Craik CS. Recombinant HIV2 protease processes HIV1 Pr53gag and analogous junction peptides in vitro. J Biol Chem. 1990;265:13890-13898. [PubMed] |
| 23. | Henderson LE, Benveniste RE, Sowder R, Copeland TD, Schultz AM, Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne). J Virol. 1988;62:2587-2595. [PubMed] |
| 24. | Battles JK, Hu MY, Rasmussen L, Tobin GJ, Gonda MA. Immunological characterization of the gag gene products of bovine immunodeficiency virus. J Virol. 1992;66:6868-6877. [PubMed] |