Published online Apr 14, 2005. doi: 10.3748/wjg.v11.i14.2157
Revised: April 9, 2003
Accepted: May 19, 2003
Published online: April 14, 2005
AIM: To study the effect of hepatitis C virus nonstructural region 3 (HCV NS3) protein on proliferation and transformation of normal human liver cell line.
METHODS: QSG7701 cells were transfected with pRcHCNS3-5’, pRcHCNS3-3’ and pRcCMV using lipofectamine transfecting technique and selected with G418 method. Expression of HCV NS3 protein was determined by immunohistochemistry. Biologic characteristics of transfected cells were evaluated by population doubling time and soft agar assays. Activation of MAPK was analyzed using Western blot with phosphospecific monoclonal antibody against dually phosphorylated MAPK.
RESULTS: QSG7701 cells transfected with pRcHCNS3-5’ showed strong intracellular expression of HCVNS3 protein, and the positive signal was localized in cytoplasm. The expressing strength of HCVNS3 protein in pRcHCNS3-3’-transfected cells was weaker than that in pRcHCNS3-5’-transfected cells. The population doubling time in the transfected cells with pRcHCNS3-5’ (12 h) was much shorter than those with pRcHCNS3-3’, pRcCMV and normal cells (24, 26, 28 h, respectively) (P<0.01). The transfected cells with pRcHCNS3-5’ showed much more anchorage independent colonies than that in those with pRcHCNS3-3’ and pRcCMV (P<0.01). The cloning efficiencies of transfected cells with pRcHCNS3-5’, pRcHCNS3-3’, pRcCMV and controls were 33%, 1.33%, 1.46%, 1.11% respectively. The level of phosphorylated MAPK in the cells with pRcHCNS3-5’ was much higher than that in those with pRcHCNS3-3’and pRcCMV and normal cells (P<0.01).
CONCLUSION: The results suggest that (1) QSG7701 cells are a better human liver cell line for investigating the pathogenesis of HCV NS3 protein. (2) 5’ region of the HCV genome segment encoding HCV NS3 is involved in cell growth and cell phenotype. (3) HCV NS3 N-terminal peptide may up-regulate the activation of MAPK, but not affect the expression of MAPK.
- Citation: Feng DY, Sun Y, Cheng RX, Ouyang XM, Zheng H. Effect of hepatitis C virus nonstructural protein NS3 on proliferation and MAPK phosphorylation of normal hepatocyte line. World J Gastroenterol 2005; 11(14): 2157-2161
- URL: https://www.wjgnet.com/1007-9327/full/v11/i14/2157.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i14.2157
Hepatitis C virus nonstructural region 3 (HCV NS3) gene, located in nucleotide 3420-5312, encodes HCV NS3 protein consisted of 631 amino acids. HCV NS3 protein is one of the important pathogenic HCV proteins, which was found and researched at first. Though it was reported that HCV NS3 protein probably has many kinds of potential biological effects, for example, proteinase and helicase activity, mediating cellular immune response, transactivating telomerase, regulating p53 function, and affecting protein kinase A (PKA) and signal transducers and activators of transcription (STAT) signal transduction, etc.[1-6]. So far the precise pathogenic mechanism of HCV NS3 protein remains unclear. Sakamuro et al[7], confirmed that NIH3T3 cells could be transformed by HCV NS3 protein and formed tumors in nude mice. Because NIH3T3 strain is a mouse fibroblast cell line, and its differentiation characteristics are different from human hepatocytes, NIH3T3 cell transformation experiment cannot really reflect the carcinogenesis process of HCV infection. In view of this, human hepatocyte line QSG7701 was transfected with mammalian expression plasmid pRcHCNS3, and the effect of HCV NS3 protein on human hepatocyte transformation and mitogen-activated protein kinase (MAPK) signal transduction were studied.
The mammalian expression plasmid pRcHCNS3-5’ (nucleotides 3354-4210, expressing HCV NS3 N-terminal peptide, Ile1020-Thr1295) and pRcHCNS3-3’ (nucleotides 4116-5147, expressing HCV NS3 C-terminal peptide, Phe1263-Trp1608) were the kind gifts of Professor Takegami[7]. Non-expressive plasmid pRcCMV was purchased from Sigma Company, USA. Human hepatocyte line QSG7701 was from Cytobiology Research Institute of Shanghai. LipofectaminTM kit and G418 were the products of GIBCO (Germany). XbaI and buffer were purchased from Sino-American Biotechnique Inc. (Shanghai, PR China), anti-HCV NS3 protein MAb from Boshide (Wuhan, PR China), S-P detection kit from Maxim Biotech Inc. (Fuzhou, PR China), p42/44MAPK (Erk1/Erk2, Thr202/Tyr204) which were used to detect the phosphorylation of MAPK was presented by New England Biolab, USA, antibody to MAPK was purchased from Santa Cruz. PCR primers for amplifying HCV NS3-5’ gene were synthesized at Shanghai Sangon (Shanghai, PR China).
Untransfected QSG7701 cells, QSG7701 cells transfected with blank plasmid pRcCMV, QSG7701 cells transfected with plasmid pRcHCNS3-5’ and QSG7701 cells transfected with plasmid pRcHCNS3-3’.
QSG7701 cells were cultured and passaged in DMEM medium with 10% fetal calf serum in an incubator containing 50 mL/L CO2 at 37 °C.
Plasmids pRcCMV, pRcHCNS3-3’ and pRcHCNS3-5’ were transferred into Escherichia coli, which was dealt with calcium chloride respectively. The E.coli was cultured to amplify the three kinds of plasmid. A small amount of the plasmids was prepared from the E.coli to identify specificity of the plasmids. The plasmids pRcHCNS3-3’ and pRcHCNS3-5’ were digested with XbaI, resolved with agarose gel electrophoresis, and stained with ethidium bromide. After identification, the plasmids were massively extracted and purified for transfecting QSG7701 cells.
QSG7701 cells were transfected with plasmids pRcCMV, pRcHCNS3-3’ or pRcHCNS3-5’ respectively as described in instruction of Lipofectaminen reagent. Cells were seeded into selection medium containing G418 until G418-resistant clones were obtained. Non-transfected QSG7701 cells were used for parallel control.
To detect cDNA in stable transfectants, total genomic DNA was extracted from positive clones according to standard methods and subjected to PCR and agarose gel electrophoresis analysis. Based on published sequences, the primers, 5’-CGGGCACGTTGTAGGCATC-3’ (sense) and 5’-AACGGACGGCTTTAGGACGA-3’ (antisense), were projected for amplifying the 5’-half sequence of HCV NS3 region[7]. PCR conditions were 35 cycles of three steps (at 94 °C for 30 s, at 57 °C for 30 s, at 72 °C for 40 s) in a 50 μL reaction mixture containing 5 μL 10×buffer, 5 μL 2 mmol/L dNTPs, 0.5 μL each primer (25 pmol/μL), 1 μL DNA, 5 U Taq DNA polymerase, and 37.5 μL distilled water. PCR products were subjected to electrophoresis on a 0.8% agarose gel for 30 min (voltage: 80 V), visualized by ethidium bromide staining.
S-P method was used to detect expression of HCV NS3 protein in the QSG7701 cells transfected with plasmids pRcHCNS3-5’, pRcHCNS3-3’, pRcCMV or non-transfected. PBS was used as substitutes of Mabs for blank control groups. Hepatocellular carcinoma tissues expressing HCV NS3 protein was used as a positive control.
Survey of growth curve Non-transfected and transfected QSG7701 cells (6×104 cells per well) were inoculated and incubated in 24-well culture plates respectively, digested and counted at an interval of 24 h, and the average number of cells in 3 wells per group was counted per time. The detection was continued for 8 d. Then the population doubling time and growth curve of the cells were calculated. To get reliable results, experiments were repeated thrice.
Anchorage-independent growth test To examine the ability of G418-resistant cells to grow anchorage independently, 2×103 cells were suspended in 0.35% agarose containing DMEM and 10% fetal calf serum and overlaid onto a bottom layer of 0.7% agarose in culture plates (φ 60 mm). After 2 wk of culture, clones with more than 50 cells were scored and the colony formation efficiency was determined (colony formation efficiency = colony numbers/seeded cell numbers×100%). The experiments were repeated thrice.
Western blot was used to detect expression and activity of MAPK. Briefly, cells (5×105) forming clones were inoculated in a 6-well culture plate and cultured for 24 h. The cells were incubated in free-serum medium for 24 h to make cell synchronous, then cultured in medium with 10% FCS for 5 h to stimulate cell growth. The cells were harvested and washed twice with pre-cooled PBS. Eighty microliters of lysis reagent (62.5 mmol/L Tris-HCl, 20 g/L SDS, 10% glycerol, 50 mmol/L DTT, 1 g/L bromophenol blue) was added and broken with ultrasound for 5-7 s, boiled at 95-100 °C for 5 min, centrifugalized at 12000 g for 5 min. Twenty microliters of supernatant proteins were resolved by SDS-10% polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred onto a nitrocellulose membrane, blocked in 5% skimmed milk for 3 h, probed with the antibody to phosphorylated p44/42 MAPK (1:100) overnight at 4 °C. After being washed thrice in TBS containing 0.1% Tween 20, the membrane was treated with horseradish peroxidase-conjugated anti-mouse antibody for 1 h. Protein binding was detected by chemiluminescence reagent (ECL). Then the membranes were eluted in cleaning solution at 50 °C for 30 min, total MAPK was detected with antibody to non-phosphorylated MAPK as described above. The bands on X-film were assayed by densitometric scanning.
Analysis of variance and t test were used according to SPSS 10.0.
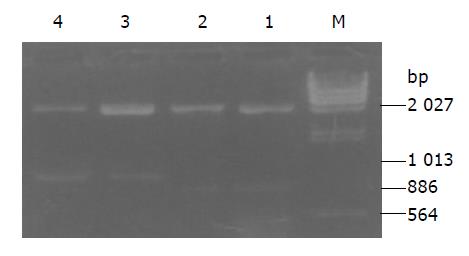
Plasmids pRcHCNS3-3’ and pRcHCNS3-5’ extracted from E.coli were digested with XbaI, resolved with agarose gel electrophoresis, and stained with ethidium bromide. As shown in Figure 1, electrophoresis analysis revealed that the major 866- and 1031-bp fragments were expected from the plasmids pRcHCNS3-3’ and pRcHCNS3-5’.
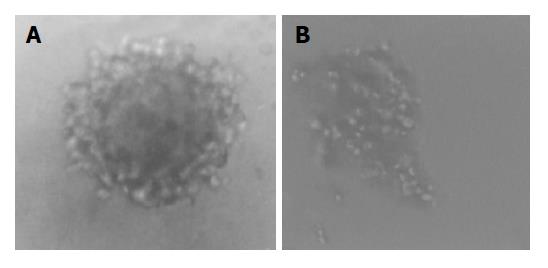
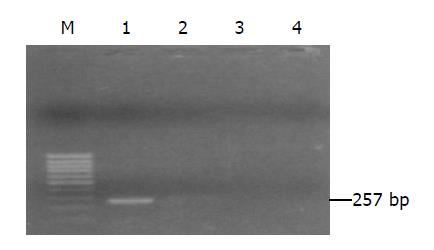
Cells transfected were seeded into selection medium containing G418. Nine pRcHCNS3-5, eight pRcHCNS3-3’ and five pRcCMV positive clones were selected (Figure 2). Total genomic DNA extracted from transfected cells was amplified by PCR. As shown in Figure 3, 257-bp fragment was specifically amplified from DNA of the QSG7701 cells transfected with plasmid pRcHCNS3-5’, and not amplified from DNA of the QSG7701 cells transfected with plasmid pRcHCNS3-3’, pRcCMV and non-transfected NIH3T3 cells.
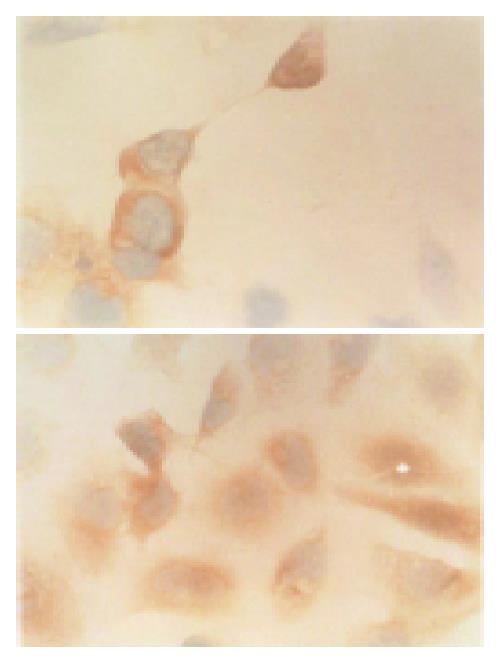
Immunohistochemical staining showed that HCV NS3 protein was expressed in transfected QSG7701 cells with plasmids pRcHCNS3-3’ and pRcHCNS3-5’. The positive signal of HCV NS3 protein was located in cytoplasm. The signal intensity of HCV NS3 protein in QSG7701 cells transfected with plasmid pRcHCNS3-5’ was higher than that in cells transfected with plasmid pRcHCNS3-3’ (Figure 4). The positive products were also found in positive control group, but not in blank and negative control groups.
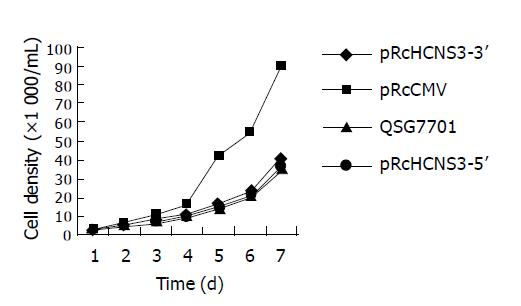
The growth curve of four kinds of cells was detected (Figure 5). The population doubling time of QSG7701 cells transfected with pRcHCN3-5’, pRcHCN3-3’, pRcCMV and non-transfected QSG7701 cells was 12, 24, 26, and 28 h, and the efficiency of colony formation was 33%, 1.33%, 1.46% and 1.11%, respectively. These showed that the population doubling time and colony formation efficiency of the cells transfected with pRcHCNS3-5’ were much shorter and higher than those of cells transfected with pRcHCNS3-3’, pRcCMV and non-transfected QSG7701 cells.
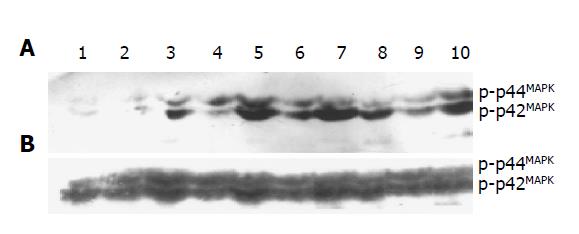
Phosphorylation of p44/42MAPK in the cells transfected with pRcHCNS3-5’, pRcHCNS3-3’, pRcCMV and non-transfected cells was detected by Western blot. The optical density of phosphorylated MAPK was 8858±887, 5612±656, 2212±245, 989±188, respectively. The level of phosphorylated p44/42MAPK in the cells transfected with pRcHCNS3-5’ was the highest (P<0.01). Expressions of total p44/42MAPK in all groups were not different (12000±1174, 11851±1048, 11321±987, 11058±991, respectively, P>0.05, Figure 6).
HCV, a kind of hepatotropic virus, causes hepatopathy by specific hepatocyte-virus reaction. Its natural host cell is hepatocyte whose differentiation characteristics are different from other cells. It is very difficult to culture and passage normal hepatocytes in vitro. Neither could hepatocytes effectively express selection markers for eukaryotic expression plasmids and obtain stable clones, nor could be transformed successfully with HCV genes. Liver cancer cell lines are usually used as a substitute of normal hepatocytes for expressing HCV gene, but they could not be used as subject cells to identify the effect of HCV genes on cell phenotype[8]. Sakamuro et al[7], reported that HCV NS3 protein could transform NIH3T3 cells, and NIH3T3 cells transfected with pRcHCN3-5’ formed tumors in nude mice. NIH3T3 cells is a mouse fibroblast line, therefore transforming NIH3T3 cells by HCV cannot really reflect the processes of infection and pathopoiesis of HCV. QSG7701 cells are an immortal normal human hepatocyte line, which were taken from liver tissue at 6 cm distance from primary liver cancer. In our experiment, QSG7701 cell line was successfully transfected with eukaryotic expression plasmids pRcHCNS3-5’, pRcHCNS3-3’and pRcCMV, and nine clones of cells transfected with pRcHCNS3-5’, eight pRcHCNS3-3’ and five pRcCMV positive clones were obtained respectively. It was identified by immunohistochemistry that all of pRcHCNS3-5’and pRcHCNS3-3’positve clones expressed HCV NS3 protein. QSG7701 cells are a kind of human normal phenotype hepatocyte strains. We first used them as subject cells to study the effect of HCV on tumorigenesis and obtained satisfactory results, and built a good cell model expressing HCV NS3 protein for studying the pathogenesis of HCC related to HCV NS3 protein.
HCV is a positive-strand RNA virus which does not have reverse transcriptase activity, and thus there is no integration of viral genome or genome segments into host chromosomes. Up to now, it has been suggested that the malignant transformation of host cells may be caused by HCV expression gene products[9,10], in which HCV NS3 protein may play an essential role in hepatocarcinogenesis, but its exact mechanism is still unclear. HCV NS3 protein containing 631 amino acids is a multifunctional cytoplasmic protein whose N-terminal has proteinase activity and C-terminal helicase activity[11]. In our data, QSG7701 cells expressing HCV N3 N-terminal peptides showed the characteristics of tumor cells to a certain extent and formed tumor in nude mice (data not shown). The population doubling time was the shortest, and anchorage-independent growth ability was the strongest. Whereas, there was no difference among growth characteristics of pRcHCNS3-3’-transfected, pRcCMV-transfected and non-transfected QSG7701 cells. The results suggest that HCVNS3 N-terminal peptide functioning as proteinase activity may play an important role during transformation of cells, which is identical with Sakamuro et al[7]. HCV NS3 protein might activate oncogenes and signaling transduction molecular of host cells infected by HCV[2,4,5]. Ras-Raf-MAPK signaling pathway is closely related to growth and proliferation of cells. MAPK could be activated by phosphorylation. Continuously high level of MAPK activity was a key for transformation and carcinogenesis of cells[12,13]. Our data displayed that HCVNS3 N-terminal peptide could significantly up-regulate phosphorylation of p44/42MAPK, but not affect the expression of total MAPK protein. Cells expressing HCVNS3 N-terminal peptide showed significant transformation phenotype, suggesting that HCV NS3 protein may induce and promote cell transformation by activating MAPK signaling pathway.
The precise mechanism of up-regulating MAPK phosphorylation is still unclear, but it has been reported that HCV NS3 protein interfered phosphorylation of proteins and inhibited cAMP-dependent PAK signaling transduction[4]. In Ras-Raf-MAPK cascade, the interaction between Ras and Raf contact points is essential for the plasma membrane localization of Raf, which ultimately leads to kinase activation. The formation of this protein complex is negatively regulated by PKA through phosphorylation of the c-Raf-1 N-terminus. Phosphorylation of c-Raf-1 serine 43 is believed to cause a N-terminal cap structure to cover the Ras docking site and inhibit Raf activation[14-17]. HCV NS3 protein (1487-1500) contains a arginine-rich sequence which is highly homologous to substrate recognition site of PKA R subunit, and could mediate binding of HCV NS3 protein to PKA C subunit and result in inactivation of PKA[18]. HCV NS3 protein may activate Ras-Raf-MAPK signaling pathway through inhibiting negative regulation of PKA. Our results showed that the effect of HCVNS3 N-terminal peptide on up-regulation of MAPK activity was more significant than that of C-terminal peptide. Because HCV NS3 N-terminal peptide (nucleotides 3354-4210, Ile1020-Thr1295) does not contain the arginine-rich sequence (1487-1500), HCV NS3 protein may activate MAPK through other ways. It was reported that HCVNS3 serine protease (nucleotides 3356-4080) could induce transformation of rat fibroblasts and tumor formation in nude mice. Their experiments suggested that the transformation and tumorigenesis induced by HCV NS3 serine protease were dependent on an active enzyme[19]. In conclusion, HCV NS3 N-terminal peptide expressed by QSG7701 cells transfected with pRcHCNS3-5’ contains the sequence coding HCV NS3 serine protease. It may need further investigation whether MAPK can be activated by serine protease of HCV NS3 N-terminal peptide, and result in proliferation and transformation of hepatocytes.
| 1. | Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152-6158. [PubMed] |
| 2. | Ishido S, Muramatsu S, Fujita T, Iwanaga Y, Tong WY, Katayama Y, Itoh M, Hotta H. Wild-type, but not mutant-type, p53 enhances nuclear accumulation of the NS3 protein of hepatitis C virus. Biochem Biophys Res Commun. 1997;230:431-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011-6019. [PubMed] |
| 4. | Borowski P, Oehlmann K, Heiland M, Laufs R. Nonstructural protein 3 of hepatitis C virus blocks the distribution of the free catalytic subunit of cyclic AMP-dependent protein kinase. J Virol. 1997;71:2838-2843. [PubMed] |
| 5. | Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469-8475. [PubMed] |
| 6. | Feng D, Cheng R, Ouyang X, Zheng H, Tsutomu T. Hepatitis C virus nonstructural protein NS(3) and telomerase activity. Chin Med J (Engl). 2002;115:597-602. [PubMed] |
| 7. | Sakamuro D, Furukawa T, Takegami T. Hepatitis C virus nonstructural protein NS3 transforms NIH 3T3 cells. J Virol. 1995;69:3893-3896. [PubMed] |
| 8. | Kim DW, Harada T, Saito I, Miyamura T. An efficient expression vector for stable expression in human liver cells. Gene. 1993;134:307-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Matsuura Y, Harada T, Makimura M, Sato M, Aizaki H, Suzuki T, Miyamura T. Characterization of HCV structural proteins expressed in various animal cells. Intervirology. 1994;37:114-118. [PubMed] |
| 10. | Hino O, Kajino K. Hepatitis virus-related hepatocarcinogenesis. Intervirology. 1994;37:133-135. [PubMed] |
| 11. | Shimotohno K. Hepatitis C virus as a causative agent of hepatocellular carcinoma. Intervirology. 1995;38:162-169. [PubMed] |
| 12. | Okazaki K, Sagata N. MAP kinase activation is essential for oncogenic transformation of NIH3T3 cells by Mos. Oncogene. 1995;10:1149-1157. [PubMed] |
| 13. | Ito Y, Sasaki Y, Horimoto M, Wada S, Tanaka Y, Kasahara A, Ueki T, Hirano T, Yamamoto H, Fujimoto J. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 291] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 14. | Borowski P, Kühl R, Laufs R, Schulze zur Wiesch J, Heiland M. Identification and characterization of a histone binding site of the non-structural protein 3 of hepatitis C virus. J Clin Virol. 1999;13:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Häfner S, Adler HS, Mischak H, Janosch P, Heidecker G, Wolfman A, Pippig S, Lohse M, Ueffing M, Kolch W. Mechanism of inhibition of Raf-1 by protein kinase A. Mol Cell Biol. 1994;14:6696-6703. [PubMed] |
| 16. | Mischak H, Seitz T, Janosch P, Eulitz M, Steen H, Schellerer M, Philipp A, Kolch W. Negative regulation of Raf-1 by phosphorylation of serine 621. Mol Cell Biol. 1996;16:5409-5418. [PubMed] |
| 17. | Marshall M. Interactions between Ras and Raf: key regulatory proteins in cellular transformation. Mol Reprod Dev. 1995;42:493-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Borowski P, Heiland M, Feucht H, Laufs R. Characterisation of non-structural protein 3 of hepatitis C virus as modulator of protein phosphorylation mediated by PKA and PKC: evidences for action on the level of substrate and enzyme. Arch Virol. 1999;144:687-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Zemel R, Gerechet S, Greif H, Bachmatove L, Birk Y, Golan-Goldhirsh A, Kunin M, Berdichevsky Y, Benhar I, Tur-Kaspa R. Cell transformation induced by hepatitis C virus NS3 serine protease. J Viral Hepat. 2001;8:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |