Published online Apr 7, 2005. doi: 10.3748/wjg.v11.i13.2026
Revised: November 3, 2004
Accepted: December 9, 2004
Published online: April 7, 2005
AIM: We shall construct the small interfering RNA (siRNA) expression cassette (SEC) targeting activated K-ras gene sequence, identify more effective siRNA sequence against K-ras gene in human pancreatic cancer cell line MiaPaCa-2 by SEC and reveal the anti-cancer effects of RNA interference (RNAi) and its therapeutic possibilities.
METHODS: Three different sites of SECs were constructed by PCR. K1/siRNA, K2/siRNA and K3/siRNA are located at sites 194, 491 and 327, respectively. They were transfected into MiaPaCa-2 cells by liposome to inhibit the expression of activated K-ras. In the interfering groups of sites 194 and 491, we detected the apoptosis in cells by FACS after they were incubated for 48 h, then we tested the alternation of K-ras gene in MiaPaCa-2 cells by RT-PCR immunofluorescence, respectively.
RESULTS: Introduction of the K1/siRNA and K2/siRNA against K-ras into MiaPaCa-2 cells leads to increased apoptosis, and the number of apoptotic cells is increased compared with control cells. The tests of RT-PCR immunofluorescence show the effects of inhibiting expression of activated K-ras gene by RNA interference in the K1/siRNA and K2/siRNA groups. We also find that the introduction of K3/siRNA has no effect on MiaPaCa-2 cells.
CONCLUSION: K1/siRNA and K2/siRNA can inhibit the expression of activated K-ras but K3/siRNA has no effect, demonstrating that K1/siRNA and K2/siRNA are effective sequences against K-ras gene and K3/siRNA are not. We conclude that specific siRNA against K-ras expression may be a powerful tool to be used therapeutically against human pancreatic cancer.
- Citation: Wang W, Wang CY, Dong JH, Chen X, Zhang M, Zhao G. Identification of effective siRNA against K-ras in human pancreatic cancer cell line MiaPaCa-2 by siRNA expression cassette. World J Gastroenterol 2005; 11(13): 2026-2031
- URL: https://www.wjgnet.com/1007-9327/full/v11/i13/2026.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i13.2026
Alteration of oncogenes and tumor suppressor genes such as K-ras and p53 was highly specific for diagnosing pancreatic cancer. K-ras is involved in transducing growth-promoting signals. Point mutations of K-ras have been found in 80-100% of pancreatic cancer, suggesting that mutant K-ras is a sensitive marker for detection of pancreatic cancer[1]. RNA interference (RNAi) represents an evolutionarily cellular mechanism for controlling the expression of genes in almost eukaryotes, including humans[2]. RNAi is triggered by double-stranded RNA and results in sequence-specific degradation of homologous single-stranded target RNAs[3,4]. The mediators of mRNA degradation are small interfering RNAs (siRNAs)[5], which are produced from longer dsRNA by enzymatic cleavage in the cell. These short RNA duplexes are approximately 21 nucleotides in length and have a base-paired structure with 2-nt 3’overhangs[6]. Although discovered only recently, siRNAs have revolutionized the analysis of mammalian gene function in cultured cells. siRNA is becoming a valuable tool for target validation beyond classical tissue culture cell lines.
We report here that oncogenic alleles of K-ras can be specifically and stably inactivated in human cancer through the use of RNA interference sequences, leading to loss of tumorigenicity. Three different sites of siRNA expression cassettes (SECs) were constructed by PCR. The K1/siRNA, K2/siRNA and K3/siRNA locate the site 194, 491 and 327, respectively. They were transfected into human pancreatic cancer cell line MiaPaCa-2 by liposome to identify which siRNA is more effective to inhibit the expression of activated K-ras, with the goal of examining the role of K-ras in human pancreatic cancer.
Pancreatic cancer cell line MiaPaCa-2 was obtained from American Type Culture Collection (CRL 1420), the gift of Professor. Douglas, Cancer Research Center of Boston Medical College. The cells were grown in Dubecco’s modified Eagle’s medium (DMEM) complemented with 10% fetal calf serum, and incubated at 37 °C in a humidified atmosphere with 50 mL/L CO2. The medium was changed once in every three days, and the cells were transferred into 24-well plates at a density of 3×103 cells/well one day before transfection.
The principle of designing siRNA is as follows: 1. Select targeted region from K-ras cDNA sequence beginning 50-100 nt downstream of start condon and search for 23-nt sequence motif AA(N19). 2. Target sequence should have a GC content of around 50% and avoid regions with GC content <30% or >60%. 3. Perform BLAST homology search to avoid off-target effects on other genes or sequences. We designed the K1/siRNA located in 5’UTR, at site 194; the K2/siRNA and K3/siRNA located in CDS, at sites 491 and 327, respectively.
The DNA template and upstream primer were from LineSilenceRNATM Kit produced by Allele Biotechnology Company in the USA. The principle of designing downstream primer is: 1. Choose a target region that includes A2AN17C or A2GN17T, A2AN17T as the sense sequence of the K-ras RNA. 2. To generate antisense siRNA transcript, the following primer is synthesized: 5’ CAAAAACTGTAAAAA AN17C (or GN17T,AN17T) GGTGTTTCGTCCTTTCCACAAGA 3’, the 5’ underlined base pairs are terminator, and the 3’ pairs are template matching region. 3. To generate sense siRNA transcript, the above N17 sequence is replaced with its reverse sequence. Following the principle, three different sites of downstream primers were synthesized (BioJet Biotech Co. Wuhan) as follows:
sense primer of K1/siRNA (194 site):
5’ CAAAAACTGTAAAAAACTTGTGGTAGTTGGAGCTGGTGTTTCGTCCTTTCCACAAGA 3’,
antisense primer:
5’ CAAAAACTGTAAAAAAGCTCCAACTACCACAAGTGGTGTTTCGTCCTTTCCACAAGA 3’;
sense primer of K2/siRNA (491 site):
5’ CAAAAACTGTAAAAAGGACTCTGAAGATGTACCT GGTGTTTCGTCCTTTCCACAAGA 3’,
antisense primer:
5’ CAAAAACTGTAAAAAAGGTACATCTTCAGAGTCCGGTGTTTCGTCCTTTCCACAAGA 3’;
sense primer of K3/siRNA (327 site):
5’ CAAAAACTGTAAAAAACCTGTCTCTTGGATATTCGGTGTTTCGTCCTTTCCACAAGA 3’,
antisense primer:
5’ CAAAAACTGTAAAAAGAATATCCAAGAGACAGGT GGTGTTTCGTCCTTTCCACAAGA 3’;
sense primer of negative control:
5’ CAAAAACTGTAAAAAACTGGTTGTAGTTGGAGCT GGTGTTTCGTCCTTTCCACAAGA 3’,
antisense primer:
5’ CAAAAACTGTAAAAAAGCTCCAACTACAACCAGTGGTGTTTCGTCCTTTCCACAAGA 3’.
The PCR reactions were carried out in a final reaction volume of 50 µL, it contained: Template DNA (1 ng/µL) 1 µL, 10×PCR buffer 5 µL, 4dNTP 1 µL, 1 unit of Taq polymerase (Gibco, USA), upstream primer (20 µmol/L) 1.2 µL, downstream primer (20 µmol/L) 1.2 µL, ddH2O 37.5 µL; performed 40 cycles of PCR amplification (two-step) as follows: Denature 94 °C for 30 s; anneal and extend 72 °C for 1 min 30 s. The amplified DNA fragments were fractionated by agarose 1.5% gel electrophoresis and visualized by ethidium bromide staining. The position of the RT-PCR products is indicated at the site of 250 bp.
PCR products were purified with a QIAquick PCR purification kit (QIAGEN, USA) according to manufacturer’s instructions.
Usually a 200 µL reaction can produce 10 µg DNA after purification. The purified DNA fragments were fractionated by agarose 1.5% gel electrophoresis again and visualized by ethidium bromide staining. The position of the RT-PCR products is still indicated at the site of 250 bp.
Cells were transferred into 24-well plates at a density of 3×103 cells/well one day before transfection. We used Shuttle Reagent (Roche, USA) as transfection reagent. The protocol is as follows: (1) Three microliters of shuttle Reagent were mixed thoroughly with 30 µL serum- and antibiotic-free medium incubated for 5 min at room temperature. (2) 0.5 µg DNA (PCR product) was mixed thoroughly with 30 µL serum- and antibiotic-free medium incubated for 5 min at room temperature. (3) the transfection solution was added to the DNA solution and incubated for 15 min at room temperature. (4) The mixture of transfection was added into the well drop by drop. Six wells per site were performed; we set up some controls including normal cells, added the transfection without DNA, and added nonspecific DNA sequence. (5) The cells were incubated for 24-48 h at 37 °C in a humidified atmosphere with 50 mL/L CO2, then observed them through inverted microscope and did some tests to detect the change of cells.
Cells (1×105) were harvested and washed twice using 0.01 mmol/L PBS. The cells were fixed using 4 mL of cold 70% ethanol at 4 °C for a minimum of 4 h and then washed twice with PBS. The cells were then resuspended in 500 µL of PBS, stained by adding 200 µL of propidium iodide (50 µg/mL, Sigma) along with 20 µL of RNase (1 mg/mL, Sigma) in a 37 °C water bath for 15-20 min. The apoptosis cells were determined by FACS and analyzed using CellQuest software.
We detected alternation of the activated K-ras in MiaPaCa-2 by RT-PCR with RNA interference. Total RNA was extracted using the RNeasy midi kit from Qiagen (USA) according to manufacturer’s instructions. For RT-PCR, 10 µg of total RNA were reversely transcribed using oligo-dT primer and reverse transcriptase (Gibco, USA). cDNA was synthesized for 60 min at 37 °C. Complementary DNAs for K-ras were amplified by PCR using 5 µL MgCl2 and two units of Taq polymerase. The amplification protocols for K-ras were as follows: 30 cycles at 95 °C for 5 min, 95°C for 15 s, 58 °C for 20 s, 72 °C for 30 s and a final extension time of 6 min at 72 °C. The PCR primers were designed based on the published human K-ras cDNA sequences: sense 5’TTGAAGTGCTGTTTGGGATAA3’, antisense 5’AACATTCCTAGGTCAGCGCAAC3’. As a control, actin mRNA was amplified from the used cDNAs using the following amplification protocol: 30 cycles at 94 °C for 4 min, 94 °C for 15 s, 55 °C for 20 s, 72 °C for 40 s and a final extension time of 6 min at 72 °C. The PCR primers were: sense 5’CCATCGTCCACCGCAAAT3’, antisense 5’TGCTCGCTCCAACCGACT3’. The PCR products contained in 25 µL of the reaction volume were fractionated by agarose 1.5% gel electrophoresis and visualized by ethidium bromide staining.
The cells were digested by 0.02% EDTA from culture dishes and added in coverslips with caution. Thirty microliters of 4% paraformaldehyde were added to fix the dried cells for 30 min at room temperature, and washed with dH2O for 5 min; 30 µL of primary antibody were added (rabbit anti-human K-ras, Boshide Co.) in a 1:100 dilution with PBS to the cells, incubated in a humidified chamber for 90 min at room temperature, and washed thrice with PBS for 5 min; 30 µL of second antibody were added with FITC (goat anti-rabbit IgM, Proteintech Group Inc., USA) in a 1:100 dilution with PBS, incubated in a humidified chamber for 2 h at room temperature; again washed with PBS for 5 min, and the cells were observed and recorded using fluorescence microscope.
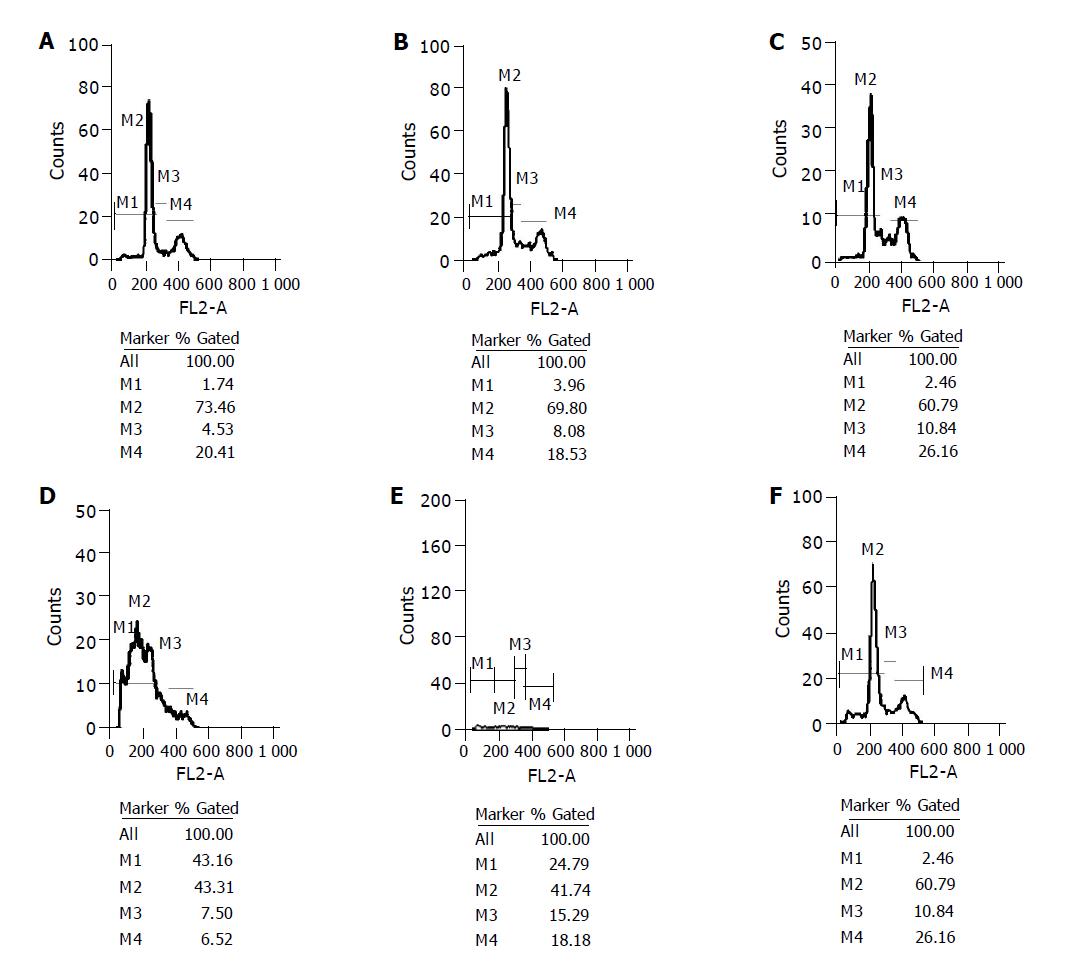
Cell cycle was determined by analysis of 1×105 cells of MiaPaCa-2 stained by propidium iodide. Cells were sorted by FACS, M1-apoptotic cells, M2-G0/G1 cells, M3-synthetic cells, M4-G2/M cells. A critical function in K-ras-mediated transformation is the increase in resistance to apoptosis. The number of apoptotic cells was increased among MiaPaCa-2 cells infected with K1/siRNA and K2/siRNA. In quantitative terms, the percentage of apoptotic cells in K1/siRNA and K2/siRNA infected cells were 43.16% and 24.79%, respectively. The percentage of apoptotic cells in K3/siRNA and nonspecific siRNA infected cells were 5.11% and 3.96%, respectively. The percentage of apoptotic cells in normal cells and only shuttle reagent infected cells were 1.74% and 2.46%, respectively. These results provide functional evidence that oncogenic mutation in K-ras is important for cell proliferation and resistance to apoptosis (Figure 1).
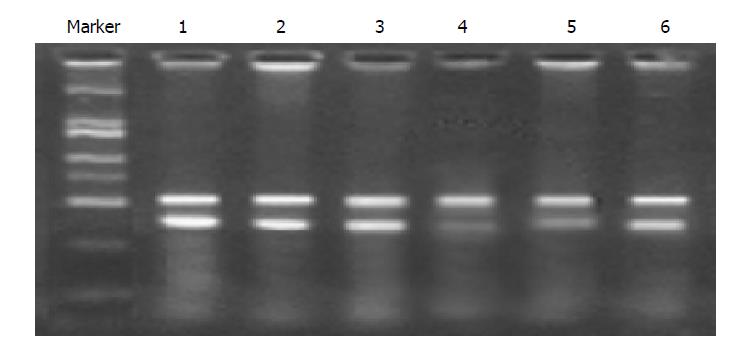
RT-PCR was used for analysis of K-ras RNA expression in MiaPaCa-2 cells. The amplified DNA fragments were fractionated by agarose 1.5% gel electrophoresis and visualized by ethidium bromide staining. It showed that the position of the RT-PCR products was located at 400 bp, and the actin product as control was located at 500 bp; the band intensity in correspondence with the cells infected with K1/siRNA and K2/siRNA became weaker compared with the control and normal cells, but the cells infected with K3/siRNA were not different from the control cells. These results provide the evidence that K1/siRNA and K2/siRNA sequences targeting K-ras can inhibit the expression of K-ras mRNA especially (Figure 2).
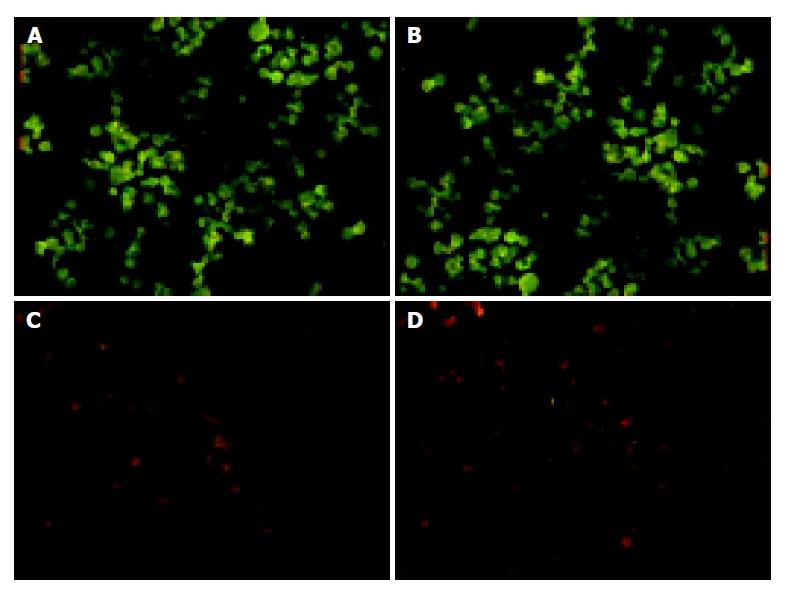
We tested whether siRNA targeting K-ras could be used to inhibit K-ras protein expression by immunofluorescence. Cells were stained with FITC antibody and observed through fluorescence microscope. The control cells displayed much green fluorescence, and it nearly could not be seen in the cells infected with K1/siRNA and K2/siRNA, but was visible in the cells infected with K3/siRNA. These results state that K1/siRNA and K2/siRNA reduce the K-ras protein expression specially in MiaPaCa-2 cells (Figure 3).
The use of PCR products to express siRNAs in mammalian cells was recently described by Castanotto et al[7]. SECs can be introduced into cells to induce gene silencing without prior cloning or sequencing. LineSilenceTM RNAi cassettes (subject of US and PCT patent application filed by Allele) are a superior SEC, which use a high level U6 RNA-based human or mouse polymerase III promoter and modified terminator, precise siRNA expression inside cells. The strategy of SECs refers to manufacturer’s description. Apparently, the activated K-ras gene may precede the development of pancreatic cancer[8,9]. It suggests that activated K-ras gene might be used in future screening protocols for pancreatic cancer and it seems to be an important target for novel anti-cancer therapies. Recently, Howard Hughes Medical Institute researcher, Jacks[10], has discovered that a gene commonly mutated in a wide range of cancers can single-handedly trigger pre-cancerous changes in cells. The discovery demonstrates that the oncogene K-ras can initiate tumor development in ways that were previously unappreciated. Therefore, we used SECs of siRNA against several sequences of the K-ras gene to selectively silence mRNA expression from activated K-ras gene in MiaPaCa-2 cells, with the goal of identifying more effective siRNA and examining the role of K-ras in human pancreatic cancer.
Brummelkamp[11] reported that they used a vector that mediated suppression of gene expression through RNAi to specifically and stably inhibit expression of only the oncogenic K-RASV12 allele in human pancreatic cancer cells. Most studies about the effect of K-ras gene in pancreatic cancer focused on codon 12 mutation of K-ras, but some pancreatic cancer cells have K-ras mutations that occur at other occasional spots or express high levels of wild-type K-ras. So we searched for different sites of siRNA sequences from human K-ras cDNA sequence, which were suitable for the principle of siRNA synthesis. We used SECs to identify the more effective siRNA sequence. We chose three sites of siRNA including site 194, 327 and 491, except for site 12. We tested cells interfered by siRNA from K-ras mRNA, also for protein expression in the cells. We draw a conclusion that different sites of siRNA had different effects on the cells. The K1/siRNA and K2/siRNA into cells led to inhibiting the expression of K-ras mRNA and protein but not K3/siRNA. The effective siRNA sequences can lead to substantial inhibition of proliferation and increased apoptotic cells, but K3/siRNA had no effect. These illustrate that activated K-ras gene plays an important role for the cell proliferation and resistance to apoptosis. As Figure 2 shows, K1/siRNA and K2/siRNA only reduced the expression of K-ras mRNA and protein but did not make it disappear, and the siRNAs were effective when they decreased the expression by over 50%[12]. The reasons why they could not inhibit expression completely are as follows: (1) Intracellular expression of the transfected PCR products had the problem of transfection efficiency; (2) Cells being in the process of persistent division, siRNA could not transfect into new born cells. K3/siRNA had no effect on the expression of K-ras and cells. It is held that some siRNA sequences designed with the exact principle were not effective. It may be associated with genome position siRNA location, which is influenced by genome secondary structure interacting with protein[7]; on the other hand, efficient RNAi requires a perfect match of 21 bases between the interfering RNA and its target sequence. Even one mismatch at certain positions could dramatically reduce the gene silencing effect[13]. We also designed control siRNA (nonspecific siRNA), whose base pairs were the same as K1/siRNA; but we changed GGTT from TGTG in K1/siRNA sequence. The PCR product of control siRNA could not influence cell proliferation; it demonstrated that the targeted gene will be knocked-down by properly designed RNAi.
In contrast to siRNA expression vectors, which require cloning and sequencing prior to use and must take 1-2 wk to prepare, SECs can be prepared in less than a day. Because they are easy to prepare, SECs are ideal for screening siRNA sequences and for identifying the most effective promoter in an experimental system. Once an effective siRNA sequence is identified, the SEC can be cloned into an appropriate vector for subsequent long-term gene silencing studies. In fact, SECs provide the perfect complement to siRNA expression vectors. The method of SECs have some deficiency, for example, it is difficult to transfect into cells, its inhibition lasts for a short while and its effect in vivo is still not clear, we had tried to use this method in vivo but failed, the data will be shown in another paper. In our experiment, we used RNAi transfection reagent, Shuttle Reagent, and QIAquick purification reagent; chose suitable dilution density of transfection and PCR products; we used serum- and antibiotic-free media as diluents; we set up several controls, including transfected shuttle reagent into cells without DNA fragment and nonspecific siRNA. What we did was to ensure that the inhibition of cell proliferation was specific. However, some problems are difficult for us, for example, how to express siRNA stably in vivo and how to make siRNA reach its effective position and its side effects on endogenous gene[14,15].
In summary, we can identify more effective siRNA sequence against K-ras gene in human pancreatic cancer cell line MiaPaCa-2 by SEC and reveal the anti-cancer effects of RNA interference. siRNA may be a powerful tool against K-ras expression, to be used therapeutically against human pancreatic cancer.
Co-correspondents: Chun-You Wang
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Minamoto T, Mai M, Ronai Z. K-ras mutation: early detection in molecular diagnosis and risk assessment of colorectal, pancreas, and lung cancers--a review. Cancer Detect Prev. 2000;24:1-12. [PubMed] |
| 2. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10136] [Article Influence: 375.4] [Reference Citation Analysis (1)] |
| 3. | Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 588] [Cited by in RCA: 578] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 4. | Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1122] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 5. | Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 189] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 6. | Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500-505. [PubMed] |
| 7. | Castanotto D, Li H, Rossi JJ. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454-1460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 106] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Lemoine NR, Jain S, Hughes CM, Staddon SL, Maillet B, Hall PA, Klöppel G. Ki-ras oncogene activation in preinvasive pancreatic cancer. Gastroenterology. 1992;102:230-236. [PubMed] |
| 9. | Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1497] [Cited by in RCA: 1454] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 10. | Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 635] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 11. | Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 843] [Cited by in RCA: 821] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 12. | Hu WY, Myers CP, Kilzer JM, Pfaff SL, Bushman FD. Inhibition of retroviral pathogenesis by RNA interference. Curr Biol. 2002;12:1301-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 128] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 13. | Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA. 2001;98:9742-9747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 737] [Cited by in RCA: 779] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 14. | Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864-5874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 337] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 15. | Haasnoot PC, Cupac D, Berkhout B. Inhibition of virus replication by RNA interference. J Biomed Sci. 2003;10:607-616. [PubMed] |