Published online Apr 7, 2005. doi: 10.3748/wjg.v11.i13.2000
Revised: October 30, 2004
Accepted: December 3, 2004
Published online: April 7, 2005
AIM: To study the effect of inhibited E-cadherin expression on invasion of cancer cells.
METHODS: We designed the nucleotide sequence of siRNA corresponding to 5’ non-coding and coding sequence of E-cadherin. 21-nucleotide dssiRNA was synthesized by in vitro transcription with Ambion Silencer TM siRNA Construction Kit. siRNA was transfected into gastric cancer MKN45 using TransMessenger transfection Kit. RT-PCR and immunofluorescent assay were used to investigate the inhibition of the expression of mutated E-cadherin. Invasive ability of cancer cells was determined by Transwell assay.
RESULTS: The synthesis of E-cadherin mRNA rather than protein expression was suppressed dramatically 7 d after interference. Decreased protein expression was observed on d 10 after interference. On d 11, invasion ability was enhanced significantly.
CONCLUSION: siRNA targeted at non-coding and coding sequence of E-cadherin showed significant inhibition on mRNA and protein expression. Inhibited E-cadherin expression results in increased invasion ability of cancer cells.
- Citation: Zheng ZH, Sun XJ, Zhou HT, Shang C, Ji H, Sun KL. Analysis of metastasis suppressing function of E-cadherin in gastric cancer cells by RNAi. World J Gastroenterol 2005; 11(13): 2000-2003
- URL: https://www.wjgnet.com/1007-9327/full/v11/i13/2000.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i13.2000
The development and metastasis of tumor are a multi-step process and involve a lot of genetic alterations. Now, many metastasis related genes were found. The recent studies indicated the loss of E-cadherin in various kinds of progressive and metastatic tumors[1]. Cell adhesion molecule, E-cadherin is capable of maintaining the polarity of epithelial cells and cell junction. It belongs to the glycoprotein family. Reduced expression of E-cadherin has been regarded as one of the main molecular events involved in dysfunction of the cell-cell adhesion system, triggering cancer invasion and metastasis[2-4]. In 1998, Guilford[5] observed three stomach cancer pedigrees of New Zealand with germline E-cadherin mutation. Thereafter, association of E-cadherin with stomach cancer received more and more attention. Recently, Brooks-Wilson et al[6], reported 41 pedigrees with genetic diffusive stomach and a pedigree with intestinal type gastric cancer. E-cadherin mutation was detected in all the 42 pedigrees. Frequency of E-cadherin mutation was lower in sporadic stomach cancer. The reduced expression of E-cadherin may be related to methylation of promoter[7]. Loss of E-cadherin expression or function was observed in many gastric cancer cell lines. Down-regulated expression of E-cadherin by both methylation of promoter and gene mutation suggested its involvement in metastasis of tumor. To further explore whether E-cadherin contributed to the metastasis inhibition, RNAi was performed to investigate changes in invasion ability of cancer cells by suppressing expression of E-cadherin in MKN45 cell line. We designed siRNA according to 5’ non-coding and coding sequence of E-cadherin and analyzed inhibition effect of siRNA. Our study will shed light on the elucidation of molecular mechanism of RNAi and its application in research of gene function and gene therapy.
Gastric cancer cell line MKN45 (poorly-differentiated adenocarcinoma) was a kind gift from Professor Yokoyama, Japan Physical and Chemical Institute. The cell line was maintained in RPMI 1640 medium supplied with 10% of FBS, 100 U/mL of penicillin and 100 µg/mL of streptomycin in a 37 °C, 50 mL/L CO2 incubator. Cells of logarithm phase were used in our study.
Primer was designed according to online tool for primer design (Ambion company: http://www.ambion.com/techlib/misc/sirna_finder.html). Twenty-nine base pair primer containing eight bases complimentary to T7 promoter and 21 to target gene was used. The following were our primers. Primer I: 5’-AACTGCAAAGCACCTGTGAGCCCTGTCTC-3’ (antisense), 5’-AAGCTCACAGGTGCTTTGCAGCCTGTCTC-3’ (sense), 12-32, NM 04360); II: 5’-AACGG-GAATGCAGTTGAGGATCCTGTCTC (antisense), 5’AAATCCTCAACTGCATTCCCGCCTGTCTC (sense), (834-854, NM 04360). Ambion Silencer TM siRNA Construction Kit was used to in vitro synthesize 21 bp sense and antisense RNA and then dsRNA by annealing. Control siRNA targeted at luciferase gene.
siRNA targeted at gene of interest was transfected according to manufacturer’s instruction (Qiagen, Chatsworth, CA, 1 µg per well, 6 µg for six-well plate). siRNA was concentrated through interaction with Enhanser R. siRNA was combined with 4 µL TransMessenger and to form the complex. The complex was diluted with 900 µL antibiotics-free medium and added into cells washed with PBS in six-well plate. During transfection, 24 h or longer after passage cells were used. Cells were washed with PBS, 2 h after transfection and grown in normal medium. mRNA inhibiting level was analyzed with RT-PCR on the seventh and the 10th d and that of protein expression with immunofluorescent cytochemistry on the 7th, 10th and the 12th d. Invasive ability of cells was analyzed with Transwell on the 11th d.
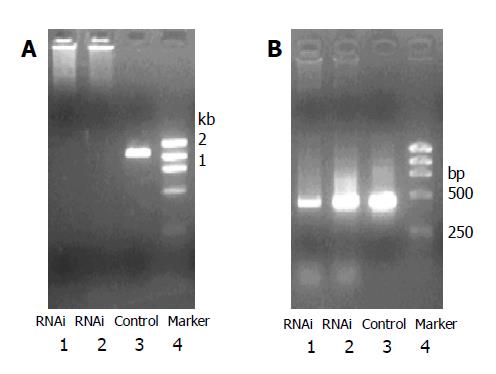
TRIzol (GIBCO BRL) was used to extract total RNA. RNA was reverse-transcribed into cDNA with reverse-transcription kit (Promega). Primers for E-cadherin were the following: upstream 5’-GGTTATTCCTCCCATCAGCT-3’, downstream: 5’-CAGTGTCCGGATTAATCTCC-3’, PCR product was 1.15 kb in length. β-actin was selected as control. Amplification fragment for β-actin was 497 bp in length. PCR conditions were the following: 95 °C for 5 min, 30 cycles of 94 °C for 30 s, 54 °C for 1 min, 72 °C 30 s, and 72 °C for 7 min. PCR products were detected by agarose electrophoresis.
MNK45 cells grown in cover slip were fixed with methanol six and 11 d after transfection and reserved in a -20 °C-refrigerator overnight. Then slips were washed with PBS followed by fixation for 6 min with acetone. Slips were washed with PBS and blocked for half an hour with non-specific serum. The first antibody (mouse anti-E-cadherin Monoclonal antibody purchased from Maixin company, Fujian) incubation continued for 2 h followed by washing with PBS. Fluorescent goat anti-mouse antibody (Zhongshan Company) was used in detection. Slips were washed with PBS. Nucleus was stained with PI for 5 min. Finally, slips were washed with PBS. Mounting was performed by using quenching-resistant reagent. The results were evaluated under fluorescent microscope.
Invasion of MKN-45 cells was assayed in Transwell cell culture chambers with 6.5-mm-diameter polycarbonate membrane filters containing 8-μm-pore size. The top chamber was prepared by coating the filter with 50 mg of diluted matrigel and incubated for 30 min. Fibroblast-conditioned medium, obtained from confluent NIH 3T3 cell cultures in serum-free RPMI 1640, was used as the chemoattractant and added to the lower wells of the chambers. The cells 10 d after interference were plated in the upper layer of costar, 2×105 cells/well. Each assay was performed thrice in triplicate. After 24 h of incubation at 37 °C, the non-migrating cells were removed from the upper surface of the membrane with a cotton swab. The cells on the lower surface of the membrane were fixed with ice-cold methanol and then stained with hematoxylin and eosin. The number of migrated cells was counted under a light microscope. Five random microscopic fields (×400) were counted per well and the mean was determined. Membrane was removed from the Traswell by cutting with a scalpel and mounted with 50% of glycerol. Five random visual fields (200×) were selected to count cells penetrating the basal membrane. Invasive ability was denoted as the average number of penetrating cells. t test was used in statistical analysis.
RT-PCR results showed significant inhibition of mRNA expression of E-cadherin in RNAi group compared with control group. siRNA targeted at upstream non-coding and coding sequence markedly inhibited expression of mutated mRNA (Figure 1).
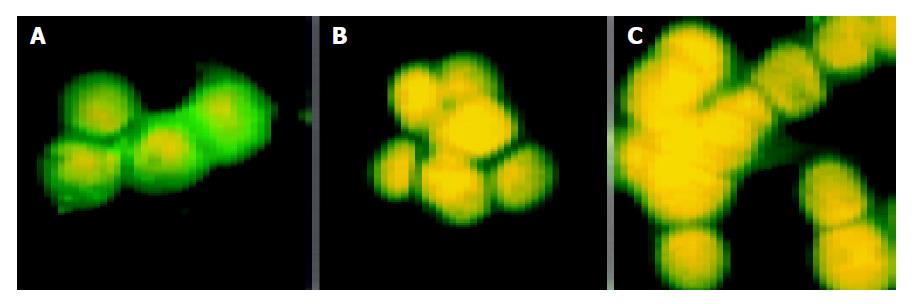
On the 7th d of interference, there was no difference in expression of E-cadherin between interference and control groups. On the 10th and 12th d of interference, intereference groups showed reduced expression of about 90% proteins suggesting inhibited expression of E-cadherin. E-cadherin was localized in cell membrane and cytoplasm and showed green fluorescence. Nucleus should show red fluorescence after PI staining. In reality, nucleus showed yellow fluorescence due to green fluorescence in cytoplasm (Figure 2).
On the 11th d of interference, E-cadherin protein expression was downregulated and invasive ability of cancer cells was upregulated. Cells penetrating filter membrane were observed before (51±6) and after interference (78±8).The difference between two groups was significant (P<0.05).
Human E-cadherin contains 882 amino acids (120 ku) and its gene maps on chromosome 16q22.1. E-cadherin protein belongs to calcium-dependent transmembrane glycoprotein mediating cell adhesion and is extensively distributed in epithelium. It is essential for tight junction and gap junction. Mutation study indicated the involvement of E-cadherin in human gastric cancer[5,6]. Investigation of E-cadherin expression in various types of gastric cancer indicated loss of E-cadherin was closely related to invasion of tumor[8,9]. In our previous study, we analyzed expression of E-cadherin in gastric cancer of different stages and found it was dramatically downregulated in progressive gastric cancer, which may be correlated with methylation of promoter[10]. It was reported that loss of E-cadherin correlated with metastasis in other tumors[11]. A direct role for E-cadherin in the suppression of tumor invasion has been demonstrated by the reversion of undifferentiated, invasive tumors to a differentiated phenotype after the transfection of E-cadherin cDNA in cell culture models[12,13]. To determine metastasis-inhibiting function of E-cadherin, we suppressed the expression of E-cadherin in MKN45 cell line by RNAi technique and found enhanced metastatic ability. It suggests that E-cadherin can inhibit tumor metastasis.
Mechanism of inhibition by E-cadherin remains unknown now. The normal E-cadherin protein is of 135 ku and processed into mature protein in golgi body, which was then released into cell surface. The half-life of E-cadherin locating in cell surface was 5 h. Newly synthesized E-cadherin binds with β-catenin and r-catenin before it is transported to cell membrane. In cell membrane, β-catenin forms complex with E-cadherin, β-catenin and r-catenin to maintain the cellular polarity. Extracellular portion of E-cadherin is necessary for calcium-dependent homotype cell adhesion, hence denoted as contact inhibitor[14-16]. Alice[17] suggested that inhibition of metastasis by E-cadherin was attributed to the binding domain for β-catenin rather than adhesion by transfecting cells that express less E-cadherin and not with E-cadherin expression vectors. Gastric cancer cell line MKN45 had mutated E-cadherin gene[18] and increased expression of mRNA and protein by two-fold compared with normal cells. Eighteen bases deletion was observed in the junction of the 6th exon and 6th intron (-10 to +8). Its cDNA had 12 bases deleted and corresponding protein had four amino acids deleted. The deleted amino acids were located between the first and second repeat domain of extracellular portion. It was also observed in our study, in which we found that the invasive ability of cancer cells were enhanced after expression of mutated E-cadherin was inhibited.
RNAi (RNA interference) is the gene-silencing technique and has emerged recently. In RNAi, dsRNA degrades homologous mRNA and hence blocks the expression of corresponding gene. Although the mechanism of RNAi remains obscure, as the simple and effective gene knock-out tool, RNAi shows great value in functional genomics study and gene therapy. Now, RNAi had been used to knock out genes in many embryo and animal cell lines such as Hela, HEK293, and P19[19,20]. We effectively inhibit the transcription of abnormal E-cadherin mRNA by using RNAi technique in gastric cancer cell line. Mutated E-cadherin abnormally expressed copious protein with half-life of 5 d. So, protein levels showed no alteration on the 7th d of RNAi. On d 10, reduced protein expression was observed. Cell transfer study suggested that invasive ability was increased after protein expression was downregulated.
Site for siRNA design was the hot spot of RNAi research and every laboratory had its own scheme. Tuschl et al[21], indicated that 5’ and 3’ UTRs should be avoided in designing siRNA considering the copious protein-binding regions. These protein and translation initiation complexes will affect the combination between siRNA endonuclease complex and mRNA, hence no detectable interference. dsRNA targeted intron and promoter sequence had no interference effect in C. elegans[22]. In plant, dsRNA targeted promoter sequence showed specific inhibition of gene expression and induction of methylation of sequence of interest[23]. So, the effective application of RNAi technique in gene function study needs further study and practice. In the current study, siRNA targeting upstream non-coding and coding sequence of E-cadherin exhibited the satisfying interference effect. It suggests that dsRNA targeting upstream non-coding sequence is capable of inhibiting gene expression. However, its effect on DNA modification needs further study. With the comprehension of mechanism of RNAi, it will promote the development of functional genomics, developmental biology and gene therapy.
Science Editor Guo SY Language Editor Elsevier HK
| 1. | Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 442] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 2. | Cavallaro U, Christofori G. Multitasking in tumor progression: signaling functions of cell adhesion molecules. Ann N Y Acad Sci. 2004;1014:58-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 3. | Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 313] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 4. | Graziano F, Humar B, Guilford P. The role of the E-cadherin gene (CDH1) in diffuse gastric cancer susceptibility: from the laboratory to clinical practice. Ann Oncol. 2003;14:1705-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1135] [Cited by in RCA: 1140] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 6. | Brooks-Wilson AR, Kaurah P, Suriano G, Leach S, Senz J, Grehan N, Butterfield YS, Jeyes J, Schinas J, Bacani J. Germline E-cadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J Med Genet. 2004;41:508-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 265] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 7. | Machado JC, Oliveira C, Carvalho R, Soares P, Berx G, Caldas C, Seruca R, Carneiro F, Sobrinho-Simöes M. E-cadherin gene (CDH1) promoter methylation as the second hit in sporadic diffuse gastric carcinoma. Oncogene. 2001;20:1525-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 195] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 8. | Chen HC, Chu RY, Hsu PN, Hsu PI, Lu JY, Lai KH, Tseng HH, Chou NH, Huang MS, Tseng CJ. Loss of E-cadherin expression correlates with poor differentiation and invasion into adjacent organs in gastric adenocarcinomas. Cancer Lett. 2003;201:97-106. [PubMed] |
| 9. | Karayiannakis AJ, Syrigos KN, Chatzigianni E, Papanikolaou S, Karatzas G. E-cadherin expression as a differentiation marker in gastric cancer. Hepatogastroenterology. 1998;45:2437-2442. [PubMed] |
| 10. | 1 Zheng ZH, Sun XJ, Ma MC, Hao DM, Liu YH, Sun KL. Studies of promoter methylation status and protein expression of E-cadherin gene in associated progression stages of gastric cancer. YiChuan XueBao. 2003;30:103-108. [PubMed] |
| 11. | Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155-163. |
| 12. | Mareel MM, Behrens J, Birchmeier W, De Bruyne GK, Vleminckx K, Hoogewijs A, Fiers WC, Van Roy FM. Down-regulation of E-cadherin expression in Madin Darby canine kidney (MDCK) cells inside tumors of nude mice. Int J Cancer. 1991;47:922-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 125] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 674] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 14. | Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 2002;34:255-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 257] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 15. | Gooding JM, Yap KL, Ikura M. The cadherin-catenin complex as a focal point of cell adhesion and signalling: new insights from three-dimensional structures. Bioessays. 2004;26:497-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 16. | Gottardi CJ, Gumbiner BM. Adhesion signaling: how beta-catenin interacts with its partners. Curr Biol. 2001;11:R792-R794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 178] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 17. | Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 195] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 18. | Oda T, Kanai Y, Oyama T, Yoshiura K, Shimoyama Y, Birchmeier W, Sugimura T, Hirohashi S. E-cadherin gene mutations in human gastric carcinoma cell lines. Proc Natl Acad Sci USA. 1994;91:1858-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 217] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557-4565. [PubMed] |
| 20. | Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 399] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 21. | Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 837] [Cited by in RCA: 836] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 22. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10139] [Article Influence: 375.5] [Reference Citation Analysis (1)] |
| 23. | Stokes T. DNA-RNA-protein gang together in silence. Trends Plant Sci. 2003;8:53-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |