Published online Mar 28, 2005. doi: 10.3748/wjg.v11.i12.1837
Revised: February 4, 2004
Accepted: February 21, 2004
Published online: March 28, 2005
AIM: To screen genes differentially expressed in mouse hepatocarcinoma ascites cell line with high potential of lymphatic metastasis.
METHODS: A subtracted cDNA library of mouse hepatocarcinoma cell line with high potential of lymphatic metastatic Hca-F and its synogenetic cell line Hca-P with a low metastatic potential was constructed by suppression subtracted hybridization(SSH) method. The screened clones of the subtracted library were sequenced and GeneBank homology search was performed.
RESULTS: Fourteen differentially expressed cDNA fragments of Hca-F were obtained with two novel genes.
CONCLUSION: SSH is a useful technique to detect differentially expressed genes and an effective method to clone novel genes.
- Citation: Cui XN, Tang JW, Hou L, Song B, Li L, Liu JW. Screening differentially expressed genes in mouse hepatocarcinoma ascites cell line with high potential of lymphatic metastasis. World J Gastroenterol 2005; 11(12): 1837-1842
- URL: https://www.wjgnet.com/1007-9327/full/v11/i12/1837.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i12.1837
Recently, studies on the mechanism of tumor metastasis have stepped into a multiple gene age stage. However, metastasis related genes available nowadays show little capability of elucidating the puzzling process of metastasis, therefore, more attention should be paid to candidate genes of metastasis. Hca-F and Hca-P are a pair of synogenetic mouse hepatocarcinoma ascites cell lines, presenting a specific potential of lymphogenetic metastasis when inoculated subcutaneously in 615 mice. Hca-F shows a high metastatic potential (>80%), while Hca-P a low potential (<30%)[1]. We employed suppression subtracted hybridization (SSH) technique to identify differentially expressed genes specific for Hca-F so as to obtain candidate genes of lymphatic metastasis.
Hca-F and Hca-P were established and maintained by our laboratory[1], inbred 615-mice were provided by the Experimental Animal House of our University.
Sixty inbred 615-mice were randomly divided into two groups. Hca-F and Hca-P tumor cell lines were inoculated subcutaneously at 2×106 tumor cells of approximately 0.1 mL cell suspension into each mouse. They were terminated on the 28th d post-inoculation, the implanted tumor and its regional lymph nodes were stained with HE and examined under a microscope. The lymph node metastatic rates of Hca-F and Hca-P tumor cells were calculated.
Total RNA was isolated by TRIZOLTM (GIBCOBRL) and mRNA was extracted according to the protocol of oligotex mRNA spin column purification kit (Qiagen). The quantity and integrity of mRNA were detected by an ultraviolet spectrometer and electrophoresed on a denaturing formaldehyde agarose stained with EtBr.
Briefly, 2 μg aliquots each of poly (A+) mRNA from the tester and the pooled driver was subjected to dscDNA synthesis, and purified by passing through Chroma spin-400 columns (Clontech, USA). Each purified dscDNA was digested with Rsa I.
mRNA of Hca-F served as a tester and mRNA of Hca-P as a driver. SSH was performed between tester and driver using a PCR selectTM cDNA subtraction kit and 50×PCR enzyme kit (Clontech, Heidelberg, Germany) following the instructions of the manufacturer.
The tester cDNAs were subdivided into two equal groups and then ligated to adaptors 1 and 2R in separate ligation reactions.
Ligation efficiency analysis was performed by amplifying ligation products with G3PDH 3’ primer, PCR primer 1 and G3PDH 3’ primer, G3PDH 5’ primer, respectively and their intensity was compared.
Subtractive hybridization was performed by annealing excess driver cDNAs with each sample of adaptor-ligated tester cDNAs. The cDNAs were heat-denatured and incubated at 68 °C for 8 h. After the first hybridization, the two samples were mixed and hybridized again with freshly heat-denatured driver cDNAs for 20 h at 68 °C. Two rounds of hybridization generated a normalized population of tester-specific cDNAs with different adaptors on each end. After the ends were filled, two rounds of PCR amplification were performed to enrich desired cDNAs containing both adaptors by exponential amplification of these products[2]. The optimized cycles for the first and second PCR were 27 and 13 respectively to increase representations and to reduce the redundancy of subtracted cDNA libraries.
Secondary PCR products were used as templates for PCR amplification of human G3PDH at 18, 23, 28 and 33 cycles to assure the subtraction efficiency. PCR products were run on an 1.8% agarose gel.
Products of the secondary PCR reactions were cloned into a pT Adv vector (CLONTECH) and the resultant ligation products were then transformed into DH5 αE. coli competent cells. The bacteria were subsequently grown in 800 μL of liquid Luria-Bertani medium and incubated for 45 min at 37 °C with shaking at 150 r/min. Thereafter, the cells were plated onto agar plates containing ampicillin (50 μg/mL), 5-bromo-4-chloro-3-indoly-b-D-galactoside (x-gal; 20 μg/cm2) and iso-ploprl-b-D-thiogalactoside (IPTG; 12.1 μg/cm2), incubated overnight at 37 °C. Individual recombinant white clones were picked and grown in a single line pattern onto Luria-Bertani agar solid medium containing ampicillin and incubated at 37 °C for 6-7 h before single clones were picked from the single-line pattern agar medium and grown in Luria-Bertani liquid medium containing ampicillin overnight at 37 °C with shaking at 150 r/min.
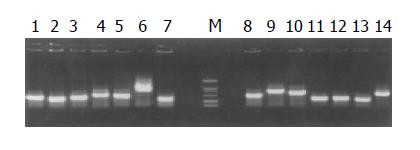
Plasmids of candidate positive clones from subtracted cDNA library were isolated and amplified by PCR with nested primer 1 and primer 2. Meanwhile the products of PCR were detected by agarose gel electrophoresis.
Fourteen positive clones from the subtracted cDNA library were sequenced by T7/SP6 chain termination reaction in TaKaRa (Dalian, China). Nucleic acid homology searches were subsequently performed at the National Center of Biotechnology Information (National Institutes of Health, Bethesda, Md., NCBI).
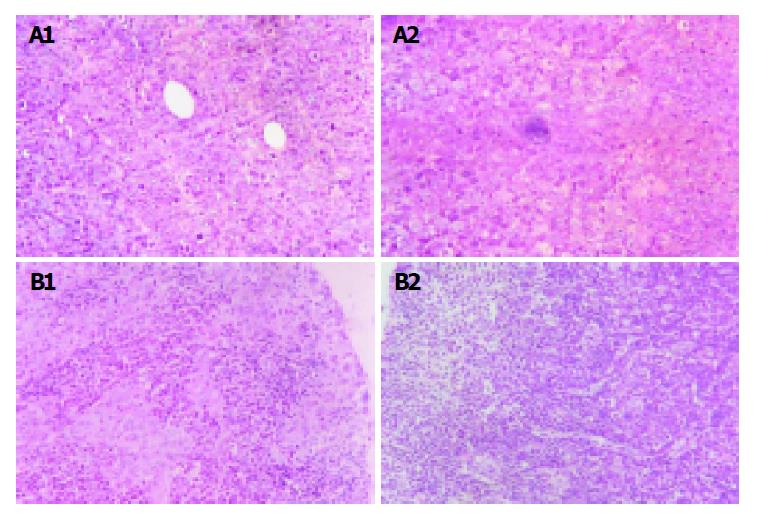
The implanted tumors both in Hca-F tumor-bearing mice and in Hca-P tumor-bearing mice were palpable on the 7th d post-inoculation.On the 28th d post-inoculation, 80% of Hca-F cell burden mice developed metastatic regional lymph node (24/30),while 10% of Hca-P cell burden mice developed metastatic regional lymph node (3/30). Figure 1A shows the implanted tumor of Hca-F cells and Hca-P cells and Figure 1B shows their regional lymph nodes.
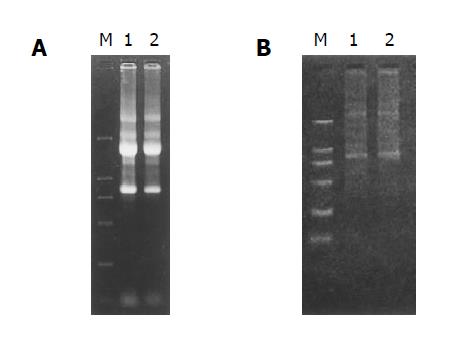
The RNA samples electrophoresed on 1% agarose/EtBr gels exhibited 2 typical bands, corresponding to ribosomal 28 s and 18 s RNA, respectively, with a ratio of intensities >2:1 and an ideal A260/A280 ratio of 1.9, indicating a high integrity and purity of the total RNA was obtained (Figure 2A).
mRNA samples appeared as a smear with a weak ribosomal RNA band, showing a high-quality mRNA was purified (Figure 2B).
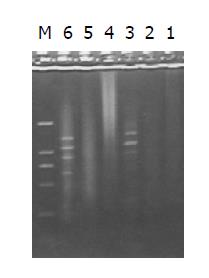
Both the digested cDNA and undigested cDNA usually looked like smears. However, the patterns among them were different, the digested cDNA fragments became short after Rsa I digestion (Figure 3).
The intensity of PCR products amplified using one gene-specific primer (G3PDH 3’ primer) and PCR primer 1 was 25%, higher than that of PCR products amplified using two gene-specific primer (G3PDH 3’ primer and 5’ primer). The ligation efficiency was higher than 25%, ensuring the tester CDNA in the following hybridization (Figure 4).
PCR products of the subtracted and unsubtracted cDNA library usually looked like smears with or without discrete bands. However, the patterns among them were different (Figure 5).
Subtraction efficiency analysis showed the effectively reduced abundance of non-differentially expressed genes. In unsubtracted cDNA library, housekeeping gene G3PDH PCR products were visible after 23 cycles of amplification and became saturated after 23-28 cycles. However, subtracted library required 33 cycles for G3PDH to be detected (Figure 6).
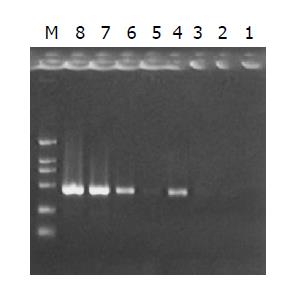
The subtracted cDNA library was composed of 995 positive clones, of which 200 clones were randomly picked up and plasmids of the candidate positive clones were isolated and amplified by PCR with nested primer 1 and primer 2. As a result, 189 positive clones showed PCR products with a length of 300-1200 bp (Figure 7).
Fourteen randomly selected clones were sequenced and homology search (http://www.ncbi.hlm.nih.gov/BLAST/) revealed 8 known genes and 3 expressed sequence tags (ESTs). Two cDNAs showed no homology and presumably represented novel genes (Table 1).
| Clone | Size (bp) | Sequence identity |
| 1-6 | 403 | Mus musculus VEGF-C |
| 10-3 | 508 | Mus musculus VEGF-C |
| 6-5 | 393 | Mus musculus ezrin |
| 22-2 | 508 | Mus musculus heat shock protein, 84 ku 1 (Hsp84-1) |
| 2-2 | 385 | Mus musculus ribosomal protein S13 (Rps13) |
| 16-1 | 274 | Mus musculus ribosomal protein L10A (Rpl10a) |
| 20-8 | 481 | Mus musculus ribosomal protein, large P2 (Rplp2) |
| 11-7 | 363 | 8 cells embryo Mus musculus cDNA clone E860014C20 |
| 40-5 | 469 | 3 cells embryo Mus musculus cDNA clone E620531C3 |
| 1-8 | 200 | EST-mouse |
| 4-5 | 405 | EST-mouse |
| 80-3 | 465 | EST-mouse |
| 1-7 | 238 | Unknown |
| 15-6 | 277 | Unknown |
As we know, mutual reaction of products of metastasis related genes could lead to tumor metastasis, therefore, elucidation of the gene expression profiles specific for tumor cells with high potential of metastasis might help search for molecular mechanisms of metastasis development. As one of the high throughput screening techniques, SSH technique is characterized by two distinct advantages. It boasts a high subtraction efficiency and harbors an equal representation of differentially expressed sequences which can separate effectively both high and low copy expressed genes mainly because of its normalization[2]. von Stein et al[3] found about 94% positive rate in their research, so they considered that confirmation of differential expression by Northern blot analysis for each clone was probably unnecessary.
A large number of studies have emphasized the angiogenesis of tumors, but the roles of lymphatic vessels in tumor growth and metastasis are neglected. However, it is well known that lymphatic metastasis is mainly responsible for the spread of epithelial malignant tumors, and its metastasis degree is closely related to the prognosis of patients.
Hca-F and Hca-P are a pair of synogenetic mouse hepatocarcinoma ascites cell lines, presenting a specific potential of lymphatic metastasis with a significant difference in their potential of metastasis[1]. Candidate genes involved in lymphatic metastasis could be obtained among differentially expressed genes.
With Hca-F as a tester and Hca-P as a driver, we used SSH technique to identify differentially expressed genes specific for Hca-F (high metastatic potential) so as to obtain candidate genes of lymphatic metastasis. As a result, 13 sequenced clones revealed 8 known genes and 3 expressed sequence tags (ESTs), 2 cDNA fragments showed no homology and presumably represented novel genes. Of the 8 known genes, VEGF-C demonstrated 2 copies. Recent studies showed that, the binding of VEGF-C to its special receptor FLT-4 on endothelial cells of stromal lymphatic vessels, could activate MAPK via intracellular kinase reaction, which could promote intratumoral lymphangiogenesis, regulate permeability of lymphatic vessels[4-9] and contribute to lymphatic metastasis[10,11]. The expression of VEGF-C in breast cancer cells could increase intratumoral lymphangiogenesis, resulting in significantly enhanced metastasis to regional lymph nodes, and the degree of tumor lymphangiogenesis is highly correlated with the extent of lymph node metastasis[12]. In colorectal cancer and gastric carcinoma, high expression of VEGF-C is significantly correlated with lymphatic invasion and lymph node metastasis[13,14,]. As a membrane-cytoskeleton linker, ezrin protein is involved in the formation of microvilli and intercellular junctions, as well as the cell motility and invasive behavior of malignant tumors[15-19] and conveys invasive phenotypes to malignant tumors[20,21]. Early studies showed that HSP was well related to metastatic potential of human esophageal squamous cell carcinoma[22], gastric carcinomas[23,24] and mice pancreatic carcinoma. The high level expression of HSP27 in human hepatocarcinoma is significantly associated with tumor progression, indicating that SSH capable of enriching metastasis related genes. Barnard et al[25] identified a cDNA clone encoding human acidic ribosomal phosphoprotein (P0) overexpressed in hepatocellular carcinoma and found that the increased P0 expression, in line with certain other ribosomal proteins, might be associated with human colorectal cancer progression. Denis et al[26] and Chadeneau et al[27] screened S13 ribosomal protein between 2 rat colon-carcinoma cell lines differing in their biological aggressiveness, which suggests that the S13 ribosomal-protein gene can be used to evaluate the growth rate of tumor cells. Kondoh et al[28] proved that high expression of S19 and LBP and low expression of HLA-I were well correlated with colon carcinoma cells with a higher malignant potential. The mechanism of up-regulated expression of ribosomal protein gene remains uncertain. In the process of tumor metastasis, synthesis of functional protein is highly activitated, enhancement of some ribosomal proteins is a prerequisite for activation of protein synthesis[30]. Continuous protein synthesis is required to maintain the entry of tumor cells into S phase and the initiation of DNA synthesis[29-31]. The data from different laboratories show that up-regulated expression of ribosomal protein genes during metastasis is not a casual incident and our study first established a correlation between ribosomal protein genes and metastasis of hepatocarcinoma. Another 2 clones showed high homology with clone E860014C20 of Mus musculus 8 cells embryo cDNA library and clone E620531C3 of Mus musculus 3 cells embryo cDNA library, respectively. Embryo genes AFP and CEA overexpressed in hepatocarcinoma and colon carcinoma indicate a possible association between embryo and tumor. The role of up-regulated expression of embryo genes in tumor metastasis needs further study. Still 2 cDNA fragments demonstrated homology with 2 EST-mouse and 2 cDNA fragments showed no homology and presumably represented novel genes[32].
In conclusion, lymphatic invasiveness of tumor cells is determined by multiple genes and co-factors with complicated cellular signal pathways. Further functional study of the candidate novel genes is needed to clarify the molecular mechanism of tumor metastasis.
| 1. | Ji Y, Ling MY, Li Y, Xie H. Effect of cell fusion on metastatic ability of mouse hepatocarcinoma cell lines. World J Gastroenterol. 1999;5:22-24. [PubMed] |
| 2. | Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 243] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 3. | von Stein OD, Thies WG, Hofmann M. A high throughput screening for rarely transcribed differentially expressed genes. Nucleic Acids Res. 1997;25:2598-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 117] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 4. | Kawakami M, Furuhata T, Kimura Y, Yamaguchi K, Hata F, Sasaki K, Hirata K. Quantification of vascular endothelial growth factor-C and its receptor-3 messenger RNA with real-time quantitative polymerase chain reaction as a predictor of lymph node metastasis in human colorectal cancer. Surgery. 2003;133:300-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 5. | Kubo H, Cao R, Brakenhielm E, Mäkinen T, Cao Y, Alitalo K. Blockade of vascular endothelial growth factor receptor-3 signaling inhibits fibroblast growth factor-2-induced lymphangiogenesis in mouse cornea. Proc Natl Acad Sci USA. 2002;99:8868-8873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 221] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 6. | Karkkainen MJ, Saaristo A, Jussila L, Karila KA, Lawrence EC, Pajusola K, Bueler H, Eichmann A, Kauppinen R, Kettunen MI. A model for gene therapy of human hereditary lymphedema. Proc Natl Acad Sci USA. 2001;98:12677-12682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 447] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 7. | Yonemura Y, Fushida S, Bando E, Kinoshita K, Miwa K, Endo Y, Sugiyama K, Partanen T, Yamamoto H, Sasaki T. Lymphangiogenesis and the vascular endothelial growth factor receptor (VEGFR)-3 in gastric cancer. Eur J Cancer. 2001;37:918-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 87] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Pepper MS. Lymphangiogenesis and tumor metastasis: myth or reality? Clin Cancer Res. 2001;7:462-468. [PubMed] |
| 9. | Karpanen T, Egeblad M, Karkkainen MJ, Kubo H, Ylä-Herttuala S, Jäättelä M, Alitalo K. Vascular endothelial growth factor C promotes tumor lymphangiogenesis and intralymphatic tumor growth. Cancer Res. 2001;61:1786-1790. [PubMed] |
| 10. | Karkkainen MJ, Petrova TV. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene. 2000;19:5598-5605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 291] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 11. | Arinaga M, Noguchi T, Takeno S, Chujo M, Miura T, Uchida Y. Clinical significance of vascular endothelial growth factor C and vascular endothelial growth factor receptor 3 in patients with nonsmall cell lung carcinoma. Cancer. 2003;97:457-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 124] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 12. | Skobe M, Hawighorst T, Jackson DG, Prevo R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K, Detmar M. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat Med. 2001;7:192-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1291] [Cited by in RCA: 1298] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 13. | Akagi K, Ikeda Y, Miyazaki M, Abe T, Kinoshita J, Maehara Y, Sugimachi K. Vascular endothelial growth factor-C (VEGF-C) expression in human colorectal cancer tissues. Br J Cancer. 2000;83:887-891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 14. | Kabashima A, Maehara Y, Kakeji Y, Sugimachi K. Overexpression of vascular endothelial growth factor C is related to lymphogenous metastasis in early gastric carcinoma. Oncology. 2001;60:146-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics: Authors' correction. Trends Cell Biol. 1999;9:289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 277] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 16. | Scherer SS, Xu T, Crino P, Arroyo EJ, Gutmann DH. Ezrin, radixin, and moesin are components of Schwann cell microvilli. J Neurosci Res. 2001;65:150-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Ohtani K, Sakamoto H, Rutherford T, Chen Z, Satoh K, Naftolin F. Ezrin, a membrane-cytoskeletal linking protein, is involved in the process of invasion of endometrial cancer cells. Cancer Lett. 1999;147:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Mäkitie T, Carpén O, Vaheri A, Kivelä T. Ezrin as a prognostic indicator and its relationship to tumor characteristics in uveal malignant melanoma. Invest Ophthalmol Vis Sci. 2001;42:2442-2449. [PubMed] |
| 19. | Gautreau A, Poullet P, Louvard D, Arpin M. Ezrin, a plasma membrane-microfilament linker, signals cell survival through the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:7300-7305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 258] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 20. | Geiger KD, Stoldt P, Schlote W, Derouiche A. Ezrin immunoreactivity is associated with increasing malignancy of astrocytic tumors but is absent in oligodendrogliomas. Am J Pathol. 2000;157:1785-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Akisawa N, Nishimori I, Iwamura T, Onishi S, Hollingsworth MA. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem Biophys Res Commun. 1999;258:395-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 101] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 22. | Noguchi T, Takeno S, Shibata T, Uchida Y, Yokoyama S, Müller W. Expression of heat shock protein 70 in grossly resected esophageal squamous cell carcinoma. Ann Thorac Surg. 2002;74:222-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Kapranos N, Kominea A, Konstantinopoulos PA, Savva S, Artelaris S, Vandoros G, Sotiropoulou-Bonikou G, Papavassiliou AG. Expression of the 27-kDa heat shock protein (HSP27) in gastric carcinomas and adjacent normal, metaplastic, and dysplastic gastric mucosa, and its prognostic significance. J Cancer Res Clin Oncol. 2002;128:426-432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Piselli P, Vendetti S, Vismara D, Cicconi R, Poccia F, Colizzi V, Delpino A. Different expression of CD44, ICAM-1, and HSP60 on primary tumor and metastases of a human pancreatic carcinoma growing in scid mice. Anticancer Res. 2000;20:825-831. [PubMed] |
| 25. | Barnard GF, Staniunas RJ, Bao S, Mafune K, Steele GD, Gollan JL, Chen LB. Increased expression of human ribosomal phosphoprotein P0 messenger RNA in hepatocellular carcinoma and colon carcinoma. Cancer Res. 1992;52:3067-3072. [PubMed] |
| 26. | Denis MG, Chadeneau C, Lecabellec MT, LeMoullac B, LeMevel B, Meflah K, Lustenberger P. Over-expression of the S13 ribosomal protein in actively growing cells. Int J Cancer. 1993;55:275-280. [PubMed] |
| 27. | Chadéneau C, LeMoullac B, Denis MG. Cloning and analysis of the human S13 ribosomal protein cDNA. Nucleic Acids Res. 1993;21:2945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 28. | Kondoh N, Schweinfest CW, Henderson KW, Papas TS. Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 1992;52:791-796. [PubMed] |
| 29. | Brooks RF. Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell. 1977;12:311-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 229] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 30. | Thomas G, Martin-Pérez J, Siegmann M, Otto AM. The effect of serum, EGF, PGF2 alpha and insulin on S6 phosphorylation and the initiation of protein and DNA synthesis. Cell. 1982;30:235-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 292] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 31. | Woo MS, Ohta Y, Rabinovitz I, Stossel TP, Blenis J. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol Cell Biol. 2004;24:3025-3035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 150] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 32. | Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1556] [Cited by in RCA: 1276] [Article Influence: 37.5] [Reference Citation Analysis (0)] |