Published online Mar 21, 2005. doi: 10.3748/wjg.v11.i11.1690
Revised: September 1, 2004
Accepted: November 24, 2004
Published online: March 21, 2005
AIM: To describe molecules or genes interaction between hepatitis B viruses (HBV) and host, for understanding how virus’ and host’s genes and molecules are networked to form a biological system and for perceiving mechanism of HBV infection.
METHODS: The knowledge of HBV infection-related reactions was organized into various kinds of pathways with carefully drawn graphs in HBVPathDB. Pathway information is stored with relational database management system (DBMS), which is currently the most efficient way to manage large amounts of data and query is implemented with powerful Structured Query Language (SQL). The search engine is written using Personal Home Page (PHP) with SQL embedded and web retrieval interface is developed for searching with Hypertext Markup Language (HTML).
RESULTS: We present the first version of HBVPathDB, which is a HBV infection-related molecular interaction network database composed of 306 pathways with 1050 molecules involved. With carefully drawn graphs, pathway information stored in HBVPathDB can be browsed in an intuitive way. We develop an easy-to-use interface for flexible accesses to the details of database. Convenient software is implemented to query and browse the pathway information of HBVPathDB. Four search page layout options-category search, gene search, description search, unitized search-are supported by the search engine of the database. The database is freely available at http://www.bio-inf.net/HBVPathDB/HBV/.
CONCLUSION: The conventional perspective HBVPathDB have already contained a considerable amount of pathway information with HBV infection related, which is suitable for in-depth analysis of molecular interaction network of virus and host. HBVPathDB integrates pathway data-sets with convenient software for query, browsing, visualization, that provides users more opportunity to identify regulatory key molecules as potential drug targets and to explore the possible mechanism of HBV infection based on gene expression datasets.
- Citation: Zhang Y, Bo XC, Yang J, Wang SQ. HBVPathDB: A database of HBV infection-related molecular interaction network. World J Gastroenterol 2005; 11(11): 1690-1692
- URL: https://www.wjgnet.com/1007-9327/full/v11/i11/1690.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i11.1690
Worldwide spread of hepatitis B viruses (HBV) involves up to 350 million people infected, and areas of endemic infection such as China has about 120 million carriers[1-3]. Approximately 80% of the carriers have different levels of hepatocyte destruction, and most of them, transfer to serious consequences progressively: liver cirrhosis and hepatocellular carcinoma[4]. Information about HBV infection-related molecular interaction network is not only helpful to comprehend the mechanism of transient and chronic infections of the liver caused by HBV, but also instructive to find targets for developing new anti-HBV drugs.
Database has proved to be an efficient tool for storage and retrieval of information on molecular interaction network[5]. However, much of the available pathways’ databases at present time such as CSNDB (Cell Signaling Networks database) focus on general information of signal transduction and metabolic pathways, but not specific disease. Although some commercial pathways’ databases such as transpath[6] offer analysis, based on pathways, to user, they are not available freely and their data sets are limited. We develop a database about HBV infection-related molecular interaction network named HBVPathDB. The pathway information stored in HBVPathDB is composed of numerous reactions involved in the interaction between HBV and its host. To be convenient for integration in the web severs or lab severs, a web retrieval interface for HBVPathDB is also developed. A web retrieval interface with four-search mode is also developed to make the database convenient for the integration in the web server or lab server. HBVPathDB is freely available at http://www.bio-inf.net/HBVPathDB/HBV/.
Since a lot of physiological and biochemical processes have been involved in the complicated interaction network between HBV and host cells, it is naturally difficult to organize a series of pathways into the complex network. A pathway that refers to local response of network under interior regulating factor or exterior stimulating signal is an essential element of the network in which plenty of molecules interact with others directly or indirectly. In HBVPathDB, pathway is the essential record type of the database. Public databases are rich resources of original pictures of signal transduction and metabolic pathways. The reference databases on the web are shown in Table 1.
| Database | URL (uniform resource locator) | |
| HemoPDB | http://www.bioinformatics.med/ohio-state.edu/HemoPDB | |
| Cytokines online | Pathfinder encyclopedia | http://www.copewithcytokines.de |
| Cell signaling | Networks database | http://www.geo.nihs.go.jp/csndb |
| PathDB | http://www.ncgr.org/pathdb/architcture.htm | |
| Signal | Pathway database | http://www.grt.kyushu-u.ac.jp/eny-doc/spad.html |
| Biocarta | http://www.biocarta.com | |
| Gene | Ontology | http://www.geneontology.org |
| Cell signaling | Technology | http://www.cellsignal.com |
| Database of interacting | Protein | http://www.dip.doe-mbi.ucla.edu |
To enable users to browse the pathway information in an intuitive way, FreehandTM 10.0 is chosen as the tracer of pathway for HBVPathDB. Molecular mode criterion provided by BiocartaTM (download freely at http://www.biocarta.com/genes/index.asp) is also used to achieve standardization of graphical representation.
HBVPathDB is a relational database implemented using MYSQL DBMS. This allows the HBVPathDB to grow without concerns for performance issues and, more importantly, to discover relations between the data. The data are organized in the format appropriate for quick search in the database.
A web retrieval interface for HBVPathDB is developed to make the database easier to use. The search engine consists of a suite of programs written in Personal Home Page and Structured Query Language (SQL). This choice was made partly to ensure that the software would work on as many hardware configurations as possible. The front-end data input part of the interface is coded in Hypertext Markup Language. The web retrieval interface has been tested on Windows 98/NT/2000/XP and Linux via the most popular web browsers Netscape and Internet Explorer.
As of August 2004, HBVPathDB contains 306 pathways with 1050 molecules involved. These pathways can be classified into adhesion, apoptosis, cell activation, cell cycle regulation, cell migration, cell signal transduction, development, hematopoiesis, immunology, metabolism and neuroscience.
HBVPathDB allows users four search modes: (1) Category search, which allows users to query pathway by category names. The search result is a list of pathways, which belong to one category or multi-category; (2) Gene search, which allows users to query pathway by gene names (five genes at most). The search result is a list of pathways with the genes involved; (3) Description search: allows users to query pathway by imprecise description; (4) Unitized search: allows users to query pathway by category name, gene name and imprecise description. The search result is a list of pathways, which belong to the category, and include the gene, and possess the description.
Results are returned as a list of pathway term with a brief account of description. And user can explore the details of specific pathway through hyperlinks.
A pathway database is an extremely useful tool to identify regulatory key molecules as potential drug targets and discover mechanism of HBV-related diseases based on gene expression datasets. The query server of pathway information is available through HBVPathDB’s web retrieval interface, which provides users with a searchable index of pathway data and gives users opportunity to analyze differential gene expression.
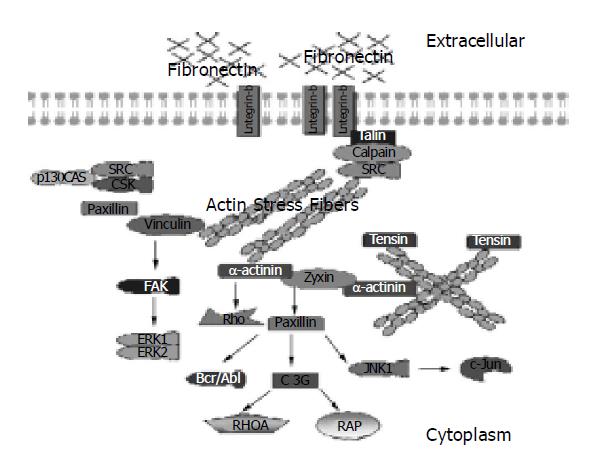
The expression of Fibronectin of hepatocyte infected with HBV is much more higher than that of normal hepatocyte in our experiment (details will be reported elsewhere). After searching for Fibronectin in HBVPathDB, we get a form with a list of five pathways associated with Fibronectin and these pathways are automatically classified into four categories: adhesion, apoptosis, cell migration and development. Click on “Integrin Signaling Pathway” in the list, then access a pathway information for which the picture is illustrated in Figure 1.
Figure 1 shows that there are a series of physiological and biochemical reactions generated after HBsAg interacted with fibronectins[7], 20 types of molecules are involved in these progresses. As a kind of cell surface receptors, integrins mediate intracellular signals in response to fibronectins, with the result that a series of pathways are activated. One of the important ultimate events shown in Figure 1 is the high expression of c-Jun gene coding a kind of DNA binding protein, which can regulate the expression of some specific genes. Furthermore, the gene of c-Jun is a type of proto-oncogene whose high expression implies carcinomatous change.
The HBVPathDB Database presented by us contains abundant information about HBV infection, ranging from HBV duplicate to cell response in host and provides user with intuitive interfaces to explore the related molecular interaction network in HBV infection process.
HBVPathDB is more than an electronic encyclopedia of HBV infection. With powerful pathway information query, navigation and visualization tools, it enables users to interpret differential gene expression generated by high-throughput technologies such as gene microarray from the point of view of pathways, and not just to find the co-express gene using clustering. The database would help to find the factors and drug targets of HBV-related diseases hidden in the complex molecular interaction network.
Science Editor Li WZ Language Editor Elsevier HK
| 1. | Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733-1745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1728] [Cited by in RCA: 1712] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 3. | Roussos A, Goritsas C, Pappas T, Spanaki M, Papadaki P, Ferti A. Prevalence of hepatitis B and C markers among refugees in Athens. World J Gastroenterol. 2003;9:993-995. [PubMed] |
| 4. | Lee YH, Yun Y. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J Biol Chem. 1998;273:25510-25515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 186] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 5. | Krishnamurthy L, Nadeau J, Ozsoyoglu G, Ozsoyoglu M, Schaeffer G, Tasan M, Xu W. Pathways database system: an integrated system for biological pathways. Bioinformatics. 2003;19:930-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Krull M, Voss N, Choi C, Pistor S, Potapov A, Wingender E. TRANSPATH: an integrated database on signal transduction and a tool for array analysis. Nucleic Acids Res. 2003;31:97-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |