Published online Mar 21, 2005. doi: 10.3748/wjg.v11.i11.1668
Revised: November 3, 2004
Accepted: November 24, 2004
Published online: March 21, 2005
AIM: Both development and progression of malignancies occur as a multistep process, requiring the activation of oncogenes and the inactivation of several tumor suppressor genes. The loss of heterozygosity (LOH) of tumor suppressor genes is believed to play a key role in carcinogenesis of colorectal cancer (CRC). In this study, we analyzed the LOH of seven loci on chromosome 22q13 in an effort to identify candidate tumor suppressor genes involved in colorectal carcinogenesis.
METHODS: Matched tumor and normal tissue DNA were analyzed by PCR using fluorescence-labeled polymorphic microsatellite markers in 83 CRC patients. PCR products were eletrophoresed and LOH was determined by calculating the peak height acquired through computer software. Comparisons between LOH frequency and clinicopathological features were performed by χ2 test. P<0.05 was considered as statistical significance.
RESULTS: The average LOH frequency of chromosome 22q13 was 28.38%. The highest LOH frequency was 64.71% on D22S1160 locus, and the lowest was 21.43% on D22S1141 locus. We detected two obvious minimal deletion regions: one between markers D22S1171 and D22S274, the other flanked by markers D22S1160 and D22S1149, each about 2.7 and 1.8 cm, respectively. None had lost in all informative loci. LOH frequency on D22S1171 is 50% on distal colon, which was higher than that on proximal one (P = 0.020); on D22S114 locus, none LOH event occurred in patients with liver metastasis, whilst 46.94% occurred in patients without liver metastasis (P = 0.008); on D22S1160 locus, LOH frequency in lymph nodes metastasis patients was 83.33%, which was much higher than 43.75% without lymph nodes metastasis ones (P = 0.016). There was no statistical significance between clinicopathological features and other loci.
CONCLUSION: This study provides evidence of two minimal deletion regions, which may harbor putative tumor suppressor genes related to progression and metastasis in sporadic colorectal carcinoma on chromosome 22q13.
- Citation: Zheng HT, Peng ZH, Zhou CZ, Li DP, Wang ZW, Qiu GQ, He L. Detailed deletion mapping of loss of heterozygosity on 22q13 in sporadic colorectal cancer. World J Gastroenterol 2005; 11(11): 1668-1672
- URL: https://www.wjgnet.com/1007-9327/full/v11/i11/1668.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i11.1668
Colorectal cancer (CRC) is a predominant disease in the western world, after lung cancer in men and breast cancer in women, and CRC is the most common cause of cancer-related death. The peak incidence of CRC was in the seventh decade of the 20th century and it is fairly equally distributed between men and women[1]. Both development and progression of malignancies occur as a multistep process, requiring the activation of oncogenes and the inactivation of several tumor suppressor genes[2]. The loss of heterozygosity (LOH) of tumor suppressor genes is believed to be one of the key steps to carcinogenesis of CRC. LOH, the loss of one allele at a specific locus, is caused by a deletion mutation or loss of a chromosome from a chromosome pair. When this occurs at a tumor suppressor gene locus where one of the alleles is already abnormal, it can result in neoplastic transformation. In CRCs, frequent allelic loss has been identified in chromosome 5q (30%), 8p (40%), 17p (75-80%), 18q (80%) and 22q (20-30%)[3]. Indeed, much has been published on tumor suppressor genes APC, p53, and DCC, which have been localized to chromosome 5q, 17p and 18q, respectively. Recently, new tumor suppressor genes, such as PTEN (10q23), FHIT (3p14), Smad4 (18q), have been found. The LOH analysis became an effective way to find informative loci and then to find candidate tumor suppressor genes. In an attempt to integrally investigate the loss of tumor suppressor genes and search for putative suppressor loci associated with tumor occurrence and progression, we have conducted a genome-wide LOH study of 83 tumor samples obtained from Chinese patients with sporadic CRC. We found that LOH frequency was higher than 35% in over 30 loci[4]. In this study, we analyzed the LOH of seven loci on chromosome 22q13 (encompassing D22S274 locus) of sporadic CRC in an effort to identify additional loci involved in colorectal tumorigenesis.
This study was based on 83 consecutively collected tumors, including 40 males and 43 females, from unrelated patients with CRC, treated at the surgical department in Shanghai No. 1 People’s Hospital, China, between 1998 and 1999. The patients’ ages ranged from 31 to 84 years with a median of 66 years. The cancerous tissue and adjacent normal tissue were frozen freshly. These tissues were cut into cubes of approximately 2 mm3 and immediately frozen in liquid nitrogen. DNA was extracted using standard methods with proteinase K digestion and phenol/chloroform purification. All patients were confirmed by pathology, and were staged by Duke’s criterion. Each patient was given his or her informed consent for the use of his or her tissues in this study.
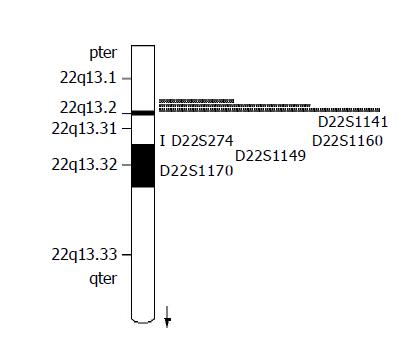
Seven fluorescence-labeled primers for polymorphic microsatellite markers (Shanghai Biological Technology Ltd, China), flanked on each side on D22S274 locus (Figure 1), were used to analyze matched pairs of normal and tumor DNA for LOH analysis. The sequence of markers was pter-D22S115-D22S1171-D22S114-D22S274-D22D1141-D22S1160-D22S1149-D22S1170-qter. The relative position was from the Genothon human genetic linkage map[5].
Polymorphic microsatellite markers were analyzed for each patient’s tumor and normal DNA by PCR (GeneAmp PCR System 9700, PE Applied Biosystems, Foster City, CA). PCR conditions were as follows: 5 µL total volume with approximately 1.4 ng of DNA as a template with 10 mmol/L standard buffer, 1.5 mmol/L Mg2+, 80 mmol/L deoxynucleotide triphosphates, 0.3 unit of Hot-start Taq polymerase and 0.06 µmol/L of each oligonucleotide primer, with the forward primer fluorescence labeled with FAM. Cycling condition consists of three stages: an initial denaturation at 96 °C for 12 min in stage I; 14 cycles each at 94 °C for 20 s, 63-56 °C for 1 min (0.5 °C decreased per cycle), 72 °C for 1 min in stage II; 35 cycles each at 94 °C for 20 s, 56 °C for 1 min, 72 °C for 1 min in stage III.
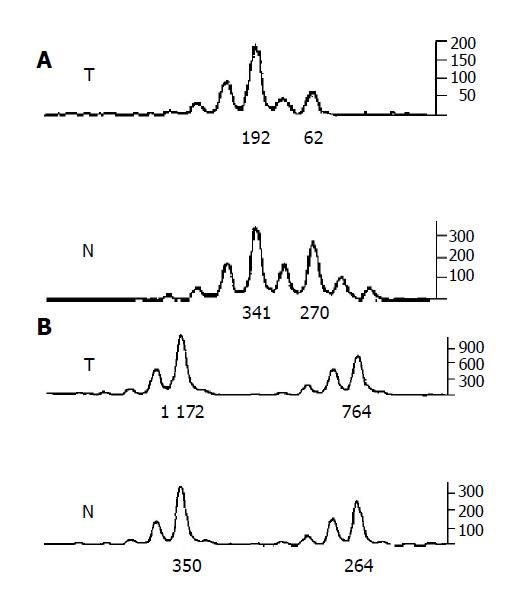
A portion of each PCR product (0.5 µL) was combined with 0.1 µL of Genescan 500 size standard (PE Applied Biosystems) and 0.9 µL of formamide loading buffer. After denaturation at 96 °C for 5 min, products were eletrophoresed on a 5% polyacrylamide gel on an ABI 377 DNA sequencer (PE Applied Biosystems) for 3 h. Genotyper 2.1 software displays individual gel lanes as electropherograms with a given size, height and area for each detected fluorescent peak. Stringent criteria were used to score the samples. Alleles were defined as the two highest peaks within the expected size range. A ratio of T1:T2/N1:N2 of less than 0.67 or greater than 1.50 was scored as a LOH (Figure 2). Most amplification of normal DNA produced two PCR products indicating heterozygosity. A single fragment amplified from normal DNA (homozygosity) and those PCR reactions, in which fragments were not clearly amplified, were scored as not informative. The LOH frequency of a locus is equal to the ratio of the number between allelic loss and informative cases. The average LOH frequency of chromosome 22 long arm is the average value of each locus LOH frequency.
Comparisons between LOH and clinicopathological data were performed by χ2 test. P<0.05 was considered as statistically significant.
The average LOH frequency of chromosome 22q13 is 28.38%. Fifty-five cases (66.27%) showed LOH on at least one marker on 22q13 (Table 1). LOH frequency on D22S1160 locus was the highest (64.71%), and on D22S1141 locus is the lowest (21.43%). We screened two obvious minimal deletion regions: one between markers D22S1171 and D22S274 (22q13.31), the other flanked by markers D22S1160 and D22S1149, each about 2.7 and 1.8 cm, respectively. None had lost in all informative loci. On D22S1149 locus, less information was got because of more homozygosity (Table 2).
| No | D22S115 | D22S1171 | D22S114 | D22S1141 | D22S1160 | D22S1149 | D22S1170 |
| 001T | △ | △ | L | △ | N | △ | N |
| 019T | N | N | △ | N | L | △ | △ |
| 002T | N | △ | N | N | △ | △ | L |
| 006T | △ | N | L | △ | △ | △ | N |
| 012T | L | N | L | △ | L | △ | △ |
| 016T | N | L | N | △ | △ | △ | △ |
| 021T | N | N | △ | N | L | △ | △ |
| 008T | △ | L | L | △ | △ | △ | △ |
| 013T | N | △ | N | N | L | △ | △ |
| 017T | N | N | L | △ | △ | △ | △ |
| 022T | N | △ | L | △ | △ | △ | △ |
| 010T | △ | N | L | △ | △ | L | N |
| 014T | △ | △ | △ | N | L | △ | L |
| 018T | N | L | △ | △ | N | △ | △ |
| 023T | △ | L | △ | N | N | N | △ |
| 029T | N | L | △ | N | L | △ | △ |
| 044T | △ | L | L | L | L | △ | △ |
| 101T | L | △ | L | △ | L | △ | L |
| 105T | N | L | △ | △ | L | △ | △ |
| 030T | △ | △ | N | L | △ | △ | L |
| 036T | N | △ | N | N | N | △ | L |
| 041T | △ | △ | L | △ | L | △ | L |
| 045T | L | L | △ | L | L | △ | N |
| 102T | △ | N | L | △ | L | △ | △ |
| 106T | L | N | △ | △ | △ | △ | △ |
| 032T | N | N | N | △ | △ | △ | L |
| 037T | N | L | △ | L | △ | △ | L |
| 042T | △ | N | N | L | △ | △ | L |
| 103T | N | △ | △ | N | L | △ | △ |
| 033T | L | L | L | L | △ | △ | L |
| 108T | N | N | N | △ | △ | △ | L |
| 116T | L | L | L | △ | L | △ | L |
| 124T | L | N | L | △ | △ | △ | △ |
| 128T | △ | △ | N | N | L | △ | △ |
| 117T | L | △ | △ | △ | △ | △ | △ |
| 121T | N | △ | N | △ | △ | L | L |
| 114T | L | N | L | △ | N | △ | L |
| 122T | L | L | L | L | △ | L | L |
| 126T | △ | △ | △ | L | L | △ | △ |
| 130T | △ | L | △ | △ | △ | L | L |
| 111T | L | L | △ | N | N | △ | L |
| 127T | △ | △ | L | △ | △ | △ | △ |
| 131T | L | N | N | △ | L | △ | △ |
| 132T | △ | L | △ | △ | L | △ | N |
| 136T | △ | N | L | △ | N | L | △ |
| 140T | N | N | N | △ | L | △ | △ |
| 144T | N | N | N | △ | △ | △ | L |
| 133T | N | △ | L | N | △ | △ | △ |
| 137T | △ | L | L | N | △ | △ | △ |
| 141T | △ | N | △ | N | L | △ | N |
| 145T | △ | N | L | N | △ | △ | △ |
| 134T | N | L | N | △ | △ | △ | △ |
| 142T | L | △ | L | L | L | N | △ |
| 139T | L | L | L | △ | △ | △ | N |
| 143T | △ | L | N | N | L | △ | △ |
| Locus | Location cases | LOH cases | Normal rate | Informative (cm) | Distance rate | LOH (%) |
| D22S115 | 22q13.2 | 14 | 36 | 60.24 | 28 | |
| D22S1171 | 22q13.31 | 19 | 31 | 60.24 | 1.2 | 38 |
| D22S114 | 22q13.31 | 23 | 35 | 69.88 | 0.7 | 39.65 |
| D22S274 | 22q13.31 | 16 | 31 | 56.63 | 2 | 34.4 |
| D22S1141 | 22q13.31 | 9 | 33 | 50.6 | 1.1 | 21.43 |
| D22S1160 | 22q13.31 | 22 | 12 | 40.96 | 0.8 | 64.71 |
| D22S1149 | 22q13.31 | 5 | 8 | 15.66 | 1.8 | 38.46 |
| D22S1170 | 22q13.31 | 7 | 28 | 42.17 | 6.8 | 20 |
LOH frequency on D22S1171 is 50% on distal large intestinal cancers, which was higher than that on proximal ones (P = 0.020). On D22S114 locus, none exhibited LOH in patients with liver metastasis, whilst 46.94% without liver metastasis (P = 0.008). On D22S1160 locus, LOH frequency in lymph metastasis patients was 83.33%, much higher than that without lymph metastasis (43.75%, P = 0.016). There was no statistical significance between clinicopathological features and other loci (Table 3).
| D22S1157 | D22S1171 | D22S114 | D22S1141 | D22S1160 | D22S1149 | D22S1170 | |||||||||
| L | N | L | N | L | N | L | N | L | N | L | N | L | N | ||
| Gender | Male | 4 | 18 | 10 | 10 | 10 | 16 | 6 | 19 | 7 | 7 | 2 | 7 | 4 | 14 |
| Female | 10 | 18 | 9 | 21 | 13 | 19 | 3 | 14 | 15 | 5 | 3 | 1 | 3 | 14 | |
| Age (yr) | >60 | 11 | 26 | 14 | 26 | 20 | 26 | 8 | 22 | 14 | 10 | 4 | 7 | 6 | 25 |
| ≤60 | 3 | 10 | 5 | 5 | 3 | 9 | 1 | 11 | 8 | 2 | 1 | 1 | 1 | 3 | |
| Location | Proximal colon | 8 | 12 | 3 | 15 | 11 | 12 | 2 | 8 | 6 | 6 | 3 | 3 | 3 | 15 |
| Distal colon | 5 | 9 | 9 | 7 | 7 | 7 | 3 | 8 | 8 | 4 | 1 | 2 | 2 | 5 | |
| Rectum | 1 | 15 | 7 | 9 | 5 | 16 | 4 | 17 | 8 | 2 | 1 | 3 | 2 | 8 | |
| Gross pattern | Massive | 7 | 14 | 7 | 13 | 11 | 14 | 4 | 14 | 5 | 7 | 4 | 4 | 4 | 11 |
| Ulcerative | 5 | 19 | 11 | 13 | 8 | 17 | 4 | 16 | 15 | 4 | 0 | 2 | 3 | 15 | |
| Encroaching | 2 | 3 | 1 | 5 | 4 | 4 | 1 | 3 | 2 | 1 | 1 | 2 | 0 | 2 | |
| Size (cm) | ≥5 | 7 | 14 | 8 | 13 | 9 | 13 | 4 | 16 | 5 | 6 | 2 | 5 | 4 | 17 |
| <5 | 7 | 22 | 11 | 18 | 14 | 22 | 5 | 17 | 17 | 6 | 3 | 3 | 3 | 11 | |
| LN metastasis | LN (+) | 7 | 20 | 10 | 17 | 12 | 17 | 4 | 16 | 15 | 3 | 2 | 3 | 3 | 13 |
| LN (-) | 7 | 16 | 9 | 14 | 11 | 18 | 5 | 17 | 7 | 9 | 3 | 5 | 4 | 15 | |
| Liver metastasis | LM (+) | 2 | 5 | 3 | 4 | 0 | 9 | 1 | 6 | 2 | 2 | 0 | 0 | 2 | 4 |
| LM (-) | 12 | 31 | 16 | 27 | 23 | 26 | 8 | 27 | 20 | 10 | 5 | 8 | 5 | 24 | |
| Differentiation | Well | 5 | 7 | 3 | 5 | 6 | 7 | 2 | 9 | 4 | 3 | 1 | 2 | 1 | 3 |
| Moderately | 6 | 23 | 13 | 15 | 10 | 21 | 6 | 15 | 10 | 6 | 2 | 3 | 6 | 16 | |
| Poorly | 1 | 1 | 1 | 3 | 2 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | |
| Mucinous | 2 | 5 | 2 | 8 | 5 | 6 | 1 | 8 | 8 | 2 | 2 | 3 | 0 | 7 | |
| Dukes stage | A | 1 | 3 | 2 | 0 | 3 | 3 | 0 | 4 | 1 | 3 | 0 | 1 | 0 | 3 |
| B | 4 | 9 | 4 | 10 | 6 | 9 | 4 | 6 | 6 | 3 | 3 | 4 | 2 | 7 | |
| C | 6 | 19 | 10 | 15 | 13 | 13 | 5 | 15 | 12 | 4 | 2 | 3 | 3 | 14 | |
| D | 3 | 5 | 3 | 5 | 1 | 10 | 0 | 8 | 3 | 2 | 0 | 0 | 2 | 4 | |
During tumorigenesis, loss of the wild-type allele is frequently observed at the appropriate locus. It was widely accepted that LOH on tumor suppressor genes played a key role in CRC transformation. LOH analysis of sporadic CRC can promote the discovery of unknown tumor suppressor genes[6,7]. Allelic loss on chromosome 22q is present not only in CRC but also in oral (40%)[6], brain (40%)[7], ovarian (55%)[8], breast (40%)[9], pancreatic endocrine tumor (30%)[10], gastrointestinal stromal tumor (77%)[11], and even hepatocellular carcinoma[12]. After microsatellite DNA analysis, several attempts were made to identify a region of deletion and eventually the tumor suppressor gene(s) responsible for these neoplasms. Allelic deletions were restricted to D22S274 (22q13) marker in oral squamous cell carcinoma[6].
We have made the LOH analysis on the long arm of chromosome 22 in the previous report[13] in CRC research, and found that there was a relatively high LOH frequency (34.04%) on D22S274 locus. In order to detect unknown tumor suppressor genes on this region, in this study, LOH scanning was carried out in 83 sporadic CRC samples with eight high-density polymorphic markers lying on each side on D22S274 locus (the average hereditary distance, 1.9 cm). By Genotyper software, i.e., by the ratio of the fluorescence intensity of allele, we hope to identify additional high-deletion loci involved in colorectal tumorigenesis and progression.
Through refined mapping, we detected two obvious minimal deletion regions: one between markers D22S1171 and D22S274 (22q13.31), LOH frequency was 34.4-39.65%, about 2.7 cm; the other flanked by marker D22S1160 and D22S1149 locus, LOH frequency was 38.46-64.71%, about 1.8 cm. Castells et al[14,15], report a allelic loss interval to a 0.5 cm on 22q13 in CRC and breast cancer. The region was flanked by D22S1171 and D22S298, which just included our first detected deletion region. Huang et al[16], found that allelic loss on 22q was 36.4% (12/33), and identified two common regions of deletion. One candidate region between D22S274 and D22S1149, about 2.4 cm, was just overlapping with our second deletion region.
LOH on D22S1171 locus was associated with tumor location, i.e., distal large intestinal cancers are prone to cause LOH than proximal cancers (P = 0.020). Zhou et al[10], found the same phenomenon on D22S274 locus. By the same result, it may be inferred that distal CRC reveals a different mechanism on tumorigenesis. Now, it is admitted that the mechanism of carcinogenesis in distal colon was different from that in proximal one[17-19]. And the mechanism in rectal cancer was also different from that in the proximal colon[20]. Distal colon cancer displayed a higher frequency of 17p and 18q allelic loss, p53 accumulation[21], c-myc expression and aneuploidy[22]. Right-sided tumors are more often diploid[23] and of the microsatellite instability (MSI) phenotype. We can conclude that LOH (17p, 18q) plays an important role in the formation of distal CRC.
Furthermore, we found that D22S1160 locus is associated with lymph nodes metastasis; 83.33% (15/18) LOH cases show lymph nodes metastasis, while only 43.75% (7/16) LOH cases without lymph nodes metastasis. This region may harbor tumor suppressor gene which associates with metastasis and results in lymph nodes invasion. But no significant difference was seen with liver metastasis. On D22S114 locus, LOH frequency is negatively associated with liver metastasis, none exhibited LOH in nine cases with liver metastasis (0/9), but 46.94% (23/49) exhibited LOH in patients without liver metastasis (P = 0.008). No significant difference was seen with lymph metastasis on this locus. These results indicate that LOH on the two loci are late events in tumorigenesis; lymph nodes metastasis and liver metastasis may reveal a different molecular mechanism.
We scanned GeneMap’ 99 database, and found that no known gene exists in these regions. Castells et al[14], also have not found known gene between D22S1171 and D22S298 locus in colorectal and breast cancer. The completion of the chromosome 22q sequencing project permitted the prediction of unknown genes using computer-based approaches. Following this strategy, the Sanger Center predicted the existence of eight genes and four pseudogenes between D22S1171 and D22S298 locus[23]. Castells and his colleague made further study and identified several DNA variants that are not compatible with pathogenic mutation. Accordingly, PARVG genes were excluded as tumor suppressor gene on 22q13 involved in CRC and breast cancer development and progression[24]. With the cloning of new genes and further function recognizing of known genes, new foundation will be achieved on 22q13.
In summary, by detailed deletion mapping, we detected two obvious LOH deletion regions, one between markers D22S1171 and D22S274 (22q13.31), the other flanked by markers D22S1160 and D22S1149, about 2.7 and 1.8 cm, respectively. These regions may harbor candidate tumor suppressor gene related to tumorigenesis and progression in CRC. Our study provided the significant data to reveal the mechanism of colorectal carcinogenesis. And further LOH scanning with high-density microsatellite markers and selected genes mutant and methylation analysis in the region may provide much more genetic and epigenetic information and find the potential tumor suppressor genes.
Science Editor Li WZ Language Editor Elsevier HK
| 1. | Ilyas M, Straub J, Tomlinson IP, Bodmer WF. Genetic pathways in colorectal and other cancers. Eur J Cancer. 1999;35:335-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 95] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 2. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8001] [Article Influence: 228.6] [Reference Citation Analysis (1)] |
| 3. | Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 879] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 4. | Peng Z, Zhang F, Zhou C, Ling Y, Bai S, Liu W, Qiu G, He L, Wang L, Wei D. Genome-wide search for loss of heterozygosity in Chinese patients with sporadic colorectal cancer. Int J Gastrointest Cancer. 2003;34:39-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 5. | Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1957] [Cited by in RCA: 1907] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 6. | Miyakawa A, Wang XL, Nakanishi H, Imai FL, Shiiba M, Miya T, Imai Y, Tanzawa H. Allelic loss on chromosome 22 in oral cancer: possibility of the existence of a tumor suppressor gene on 22q13. Int J Oncol. 1998;13:705-709. [PubMed] |
| 7. | Rubio MP, Correa KM, Ramesh V, MacCollin MM, Jacoby LB, von Deimling A, Gusella JF, Louis DN. Analysis of the neurofibromatosis 2 gene in human ependymomas and astrocytomas. Cancer Res. 1994;54:45-47. [PubMed] |
| 8. | Englefield P, Foulkes WD, Campbell IG. Loss of heterozygosity on chromosome 22 in ovarian carcinoma is distal to and is not accompanied by mutations in NF2 at 22q12. Br J Cancer. 1994;70:905-907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Iida A, Kurose K, Isobe R, Akiyama F, Sakamoto G, Yoshimoto M, Kasumi F, Nakamura Y, Emi M. Mapping of a new target region of allelic loss to a 2-cM interval at 22q13.1 in primary breast cancer. Genes Chromosomes Cancer. 1998;21:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 10. | Chung DC, Brown SB, Graeme-Cook F, Tillotson LG, Warshaw AL, Jensen RT, Arnold A. Localization of putative tumor suppressor loci by genome-wide allelotyping in human pancreatic endocrine tumors. Cancer Res. 1998;58:3706-3711. [PubMed] |
| 11. | Fukasawa T, Chong JM, Sakurai S, Koshiishi N, Ikeno R, Tanaka A, Matsumoto Y, Hayashi Y, Koike M, Fukayama M. Allelic loss of 14q and 22q, NF2 mutation, and genetic instability occur independently of c-kit mutation in gastrointestinal stromal tumor. Jpn J Cancer Res. 2000;91:1241-1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Takahashi K, Kudo J, Ishibashi H, Hirata Y, Niho Y. Frequent loss of heterozygosity on chromosome 22 in hepatocellular carcinoma. Hepatology. 1993;17:794-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Zhou CZ, Peng ZH, Zhang F, Qiu GQ, He L. Loss of heterozygosity on long arm of chromosome 22 in sporadic colorectal carcinoma. World J Gastroenterol. 2002;8:668-673. [PubMed] |
| 14. | Castells A, Gusella JF, Ramesh V, Rustgi AK. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res. 2000;60:2836-2839. [PubMed] |
| 15. | Castells A, Ino Y, Louis DN, Ramesh V, Gusella JF, Rustgi AK. Mapping of a target region of allelic loss to a 0.5-cM interval on chromosome 22q13 in human colorectal cancer. Gastroenterology. 1999;117:831-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 16. | Huang B, Starostik P, Kühl J, Tonn JC, Roggendorf W. Loss of heterozygosity on chromosome 22 in human ependymomas. Acta Neuropathol. 2002;103:415-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 550] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 18. | Distler P, Holt PR. Are right- and left-sided colon neoplasms distinct tumors? Dig Dis. 1997;15:302-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 103] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Kapiteijn E, Liefers GJ, Los LC, Kranenbarg EK, Hermans J, Tollenaar RA, Moriya Y, van de Velde CJ, van Krieken JH. Mechanisms of oncogenesis in colon versus rectal cancer. J Pathol. 2001;195:171-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Soong R, Grieu F, Robbins P, Dix B, Chen D, Parsons R, House A, Iacopetta B. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res. 1997;3:1405-1411. [PubMed] |
| 22. | Lanza G, Maestri I, Dubini A, Gafa R, Santini A, Ferretti S, Cavazzini L. p53 expression in colorectal cancer: relation to tumor type, DNA ploidy pattern and short-term survival. Am J Clin Pathol. 1996;105:604-612. [PubMed] |
| 23. | Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ. The DNA sequence of human chromosome 22. Nature. 1999;402:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 747] [Cited by in RCA: 690] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 24. | Castellví-Bel S, Castells A, Johnstone CN, Piñol V, Pellisé M, Elizalde JI, Romo N, Rustgi AK, Piqué JM. Evaluation of PARVG located on 22q13 as a candidate tumor suppressor gene for colorectal and breast cancer. Cancer Genet Cytogenet. 2003;144:80-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |