INTRODUCTION
Hereditary pancreatitis (HP; OMIM 167800) is an autosomal dominant disorder characterized by multiple episodes of acute pancreatitis, development of chronic pancreatitis and high incidence of pancreatic cancer (up to 40% of HP patients) with approximately 80% penetrance and variable expressivity[1-3]. Mutations in serine protease 1 or cationic trypsinogen (CT, PRSS1) gene have been initially identified as a causative mutation for HP[4,5]. PRSS1 encodes CT protein, which plays a crucial role in food digestion at the duodenum. It is autoactivated to trypsin by cleavage of 8 amino acids, from alanine 16 to lysine 23 residues (APFDDDDK) next to the N-terminal signal peptide of 15 residues. The activated trypsin can then autolyze itself at the primary autolysis site, arginine 122[6,7].
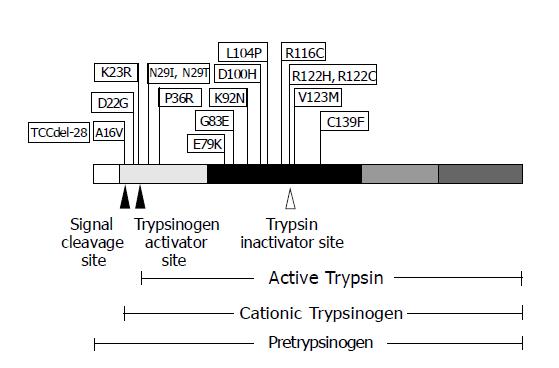
Most of the 17 PRSS1 mutations discovered, to date, occur within or near important enzymatic domains of CT and locate only in exons 2 and 3, and the promoter region of the PRSS1 gene (http://uwcmml1s.uwcm.ac.uk/uwcm/mg/search/119620.html). The majority of mutation, accounting for approximately 70% of HP cases, affects CT at arginine 122 residue by changing either to histidine (R122H) in most cases or to cysteine (R122C) in rare cases. The second most often mutation, accounting for approximately 20%, is at asparagine 29, which is frequently changed to isoleucine (N29I) and rarely altered to threonine (N29T). The two most common PRSS1 mutations (R122H and N29I) have been found in families with typical HP[4,8]. A16V and R116C are the mutations reported in several families with idiopathic chronic pancreatitis (ICP) but no history of HP with A16V having a low penetrance characteristic[9-12]. Other mutations identified in PRSS1 include TCCdel-28, D22G, K23R, P36R, E79K, G83E, K92N, D100H, L104P, V123M and C139F. However, almost all of these mutations were individually reported in a single small family without history of HP, but with ICP[11-15].
The precise mechanism by which these PRSS1 mutations cause HP disease remains unclear. However, it is believed to be a gain-of-function due to increased activity or stability of trypsin and/or decreased trypsin inactivation[7,16,17], leading to autolysis of the pancreatic tissue. Thus, the possible mechanisms may be: (1) enhanced autoactivation of trypsinogen by mutations at the autoactivation site (A16V, D22G, K23R)[9,13,15], (2) stabilization of trypsin by N29I[16], and (3) disruption of the trypsin hydrolytic recognition site by R122H[16]. A recent report by Teich et al[18] has raised a novel mechanism of action for pancreatitis-associated trypsinogen mutations. Surprisingly, E79K markedly inhibited autoactivation of CT but, in contrast, could activate anionic trypsinogen encoded by PRSS2. These findings suggested that interactions between the two major trypsinogen isoforms might also play a role in the development of HP. It has been proposed that while gain-of-function mutations of PRSS1 resulted in pancreatitis, loss-of-function mutations (Y37X and IVS2+1G>A) in the gene might provide protective effect against the disease[19].
Molecular study of HP patients has been carried out and reported mainly in Caucasian. There are limited data obtained from Eastern population, such as those obtained from Japanese, while studies of patients with chronic pancreatitis in Korea and Bangladesh could not show hot-spot mutations of PRSS1[20,21]. Here, we report a mutation of PRSS1 in a Thai family with HP and pancreatic cancer.
MATERIALS AND METHODS
Patients
A 55-year-old Thai woman was admitted to Siriraj Hospital with 3-mo history of recurrent upper abdominal pain, jaundice and steatorrhea. Biochemical testing showed diabetes mellitus, whereas cholesterol, triglyceride and HDL-cholesterol were in normal ranges. Liver enzymes were elevated (SGOT 375 U/L and SGPT 634 U/L). Diagnostic imaging demonstrated a pancreatic pseudocyst and pancreatic duct stones at the head of pancreas. Endoscopic retrograde cholangiopancreaticography was performed and showed that the first part duodenum had generalized swelling and bulging papilla. Attempt to cannulate pancreatic duct was unsuccessful and patient underwent pancreato-jejunostomy. One year after being the operation, the patient developed adenocarcinoma of the head of pancreas from which she died shortly later without blood sample collection for genetic testing. Family history revealed two additional members with confirmed history of acute pancreatitis. Patient’s son (III-2) and her younger sister (II-5) both suffered an episode of acute pancreatitis at the age of 15 and mid-30s, respectively. They are currently well without further episode, and after thoroughly examined by one of us (C.L.), no biochemical and radiographic evidence of pancreatitis could be demonstrated. Indeed, the son was the first member who became symptomatic. Moreover, the patient’s mother died in her 60 s with unconfirmed diagnosis of “abdominal cancer”. None of the symptomatic members consumed alcohol but there is a 44-year-old asymptomatic male (II-6) who had a drinking habit. There are 3 clinically symptomatic and 6 asymptomatic members of three generations in the pedigree of this family. The family consented for drawing blood in 2 affected and 6 asymptomatic members, while the proband did not give her consent before her death. Blood samples, from 54 persons who had no history of pancreatitis, were also collected as control subjects. This study was conducted in accordance with the Faculty of Medicine Siriraj Hospital Ethics Committee’s guidelines according to the Helsinki Declaration.
DNA isolation and amplification by PCR
Genomic DNA samples from peripheral blood were prepared using standard phenol-chloroform extraction procedure. Nucleotide sequence of 390 bp encompassing exon 3 of the PRSS1 gene was amplified by polymerase chain reaction (PCR) using a primer pair: PRSS1F (5’-TCCATGAGCAGAGAGCTTGAGGAA-3’) and PRSS1R (5’-TGTGAGGATGGAGGGAAGTAGAAGGACT-3’)[22]. PCR amplification was performed with 25 μL reaction volume containing 200 ng genomic DNA, 1×PCR buffer (QIAGEN, Germany), 0.2 mmol/L of each dNTP, 10 pmoL of each primer and 0.5 U Taq DNA polymerase (QIAGEN). The PCR product was generated by amplification under conditions as follows: the initial 5-min denaturation at 94 °C, followed by 35 cycles at 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s, and a final 7-min elongation step at 72 °C. Aliquot (10 μL) of the PCR reaction was then loaded onto 25 g/L agarose gel to verify the size and quantity of the PCR product prior to sequencing.
DNA sequence analysis
The PCR products from all the samples were purified using the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s protocol. Sequencing was accomplished by the use of the Applied Biosystems (ABI) Prism Big Dye Terminator Cycle Sequencing Reading Reaction Kit and ABI Prism model 310 DNA sequencer (PE Applied Biosystems, Foster City, CA). All the products were sequenced in both directions with the same forward (PRSS1F) and reverse (PRSS1R) primer pairs as used for the PCR.
Allele specific amplification
For subsequent determination of prevalence of R116C missense mutation in population, allele specific amplification (ASA) using two specifically designed primers, R116F-N (5’-ATCATGTTAATCAAGCCCTCCTGAC-3’) for the wild-type allele and C116F-M (5’-ATCATGTTAATC-AAGCCCTCCTGAT-3’) for the mutant allele, was performed in 54 normal control subjects (108 chromosomes). Each primer was included in a separate PCR mixture with a common primer, Ex3R (5’-AGCTCGTCTGGGTAGT-CGGCTGTGA-3’) to yield an allele-specific 555-bp product. An internal control fragment of growth/differentiation factor 5 (GDF5) gene was amplified by a pair of primers, GDF-3F (5’-CTGAACCCAAGCCAGGACA-3’) and GDF-3R (5’-GTACTCGTGGGGTGTGATG-3’) producing a 206-bp product. Twenty-five microliters of PCR reaction mixture contained 200 ng genomic DNA, 1×PCR buffer (QIAGEN), 0.2 mmol/L of each dNTP, 20 pmoL of each allele-specific primer, 5 pmoL of both internal control primers and 0.5 U Hot Star Taq DNA polymerase (QIAGEN). The PCR cycling condition consisted of an initial denaturation step at 95 °C for 10 min, followed by 38 cycles of 94 °C for 20 s, 53 °C for 20 s and 72 °C for 45 s, with a final elongation step at 72 °C for 10 min. The PCR products were examined by agarose gel electrophoresis using a 25 g/L agarose gel run in 0.5× TBE and stained with ethidium bromide.
RESULTS
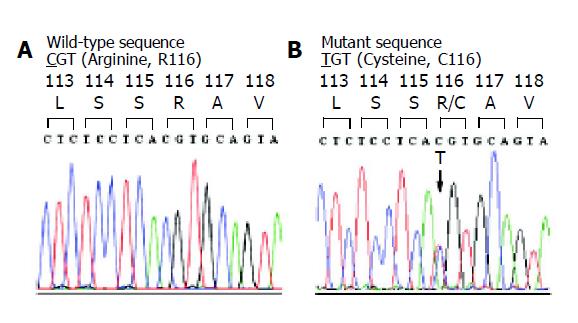
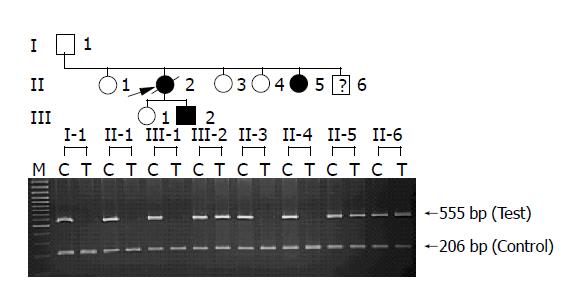
Exon 3 of PRSS1 was amplified by PCR and directly sequenced in two clinically affected members of the Thai HP family with HP. A heterozygous nucleotide substitution at position 2441 (g.2441 C>T) was identified (Figure 1), resulting in an amino acid substitution of arginine (CGT) by cysteine (TGT) at residue 116 (R116C) of the encoded protein. ASA was then developed to specifically detect the mutation in all available 8 family members as well as 54 normal control subjects. The results of ASA analysis in the HP family (Figure 2) showed the presence of R116C mutation in 3 individuals, while screening in normal control subjects did not identify the mutation (data not shown).
Figure 1 Chromatogram of DNA sequences in exon 3 of PRSS1 gene in a normal control (A) and a patient with chronic pancreatitis (B).
The tracing of the patient reveals a heterozygous missense mutation, g.2441C>T or c.346 C>T, which would result in a substitution of arginine (CGT) by cysteine (TGT) at amino acid residue 116 (R116C).
Figure 2 Allele specific amplification (ASA) analysis of R116C in the HP family.
In two affected (II-5 and III-2) and one unaffected (II-6) individuals, two bands (555 bp) amplified from both wild-type (C) and mutant (T) alleles are shown as heterozygous genotype, whereas only wild-type alleles (C) are present in the rest of unaffected individuals (I-1, II-1, II-3, II-4 and III-1). PCR products of 206 bp are shown as internal control in all lanes.
In the 3 mutation-positive individuals, 2 individuals (sister and son of the proband) had history of acute pancreatitis and 1 individual (younger brother of the proband) remained asymptomatic with an alcohol drinking habit.
DISCUSSION
CT is a trypsin precursor peptide that is synthesized within human pancreatic acinar cells and auto-activated to be trypsin by removing its signaling peptide portion of 23 amino acids. HP has been hypothesized to be caused by increased trypsin activity and/or decreased trypsin inactivation, leading to excessive trypsin in the pancreas and, ultimately, pancreatic autolysis. Mutations in PRSS1 have been initially identified as the cause of this disease since 1996[4]. To date, a total of 17 PRSS1 mutations that cause chronic pancreatitis have been described and reported (Figure 3). R122H is the first mutation identified by Whitcomb et al[4], and has been most frequently identified in more than 100 unrelated patients worldwide. N29I is the second most frequent mutation identified in more than 50 unrelated patients[8]. Comparison of patients with the two most common mutations has shown that the patients with R122H have younger age at onset and more requirements of surgical interventions than those with N29I[23,24].
Figure 3 The schematic diagram of PRSS1 mutations discovered, to date, and their positions on the encoded protein.
PRSS1 protein is shown with five regions, each of which encoded by a different exon, depicted with varying shaded blocks. The arrowheads represent the proteolysis cleavage sites of the protein resulting in various forms of trypsin biogenesis.
This study was initially aimed to analyze the two most common mutations, R122H and N29I, of PRSS1 gene in a Thai family with clear features of HP. The expected mutations were not found in the affected subjects. However, another missense mutation causing a replacement of arginine (CGT) with cysteine (TGT) at the 116th residue of CT, R116C, was identified. This mutation was initially reported as an allelic variant to GenBank as TRYP1-R116C allele (accession number AF315310). Screening of 54 normal control subjects (108 chromosomes) revealed no such change, supporting its pathogenicity. In addition, R116C is a known mutation previously reported on 3 occasions in a total of 8 patients: in two unrelated ICP patients from France[10]; in a Turkish family affected by atypical HP (only two of four members who carried this mutation in one allele developed ICP)[11]; and in a German family whose autosomal dominant inheritance of chronic pancreatitis was not clear (R116C was detected in unaffected mother and her affected young daughter)[12]. These data have demonstrated that R116C might lead to chronic pancreatitis with only low penetrance. In the present work, R116C was found in two symptomatic individuals (III-2 and II-5) and one asymptomatic member (II-6) of a Thai family. It may be possible that the disease in the latter member has not been developed or its penetrance is incomplete.
We believe that R116C occurs by deamination of cytosine in CpG dinucleotide, a well-known phenomenon that causes the C>T change. An argument can be made about the true pathogenicity of R116C since the arginine residue at position 116 is not conserved among species. However, as it has been shown from a recent computational analysis of mutational spectrum of 436 genes in the human genome, pathogenic mutations can indeed occur in non-conserved region of the gene[25]. Moreover, the nature of amino acid alteration in R116C (from non-sulfhydryl to sulfhydryl one) is considered to be far more radical than what usually observed among species[26]. Therefore, R116C is capable of being pathogenic from a functional point of view.
The mechanism of how R116C causes pancreatitis remains unclear. Computer modeling by Tautermann and colleagues[11] demonstrated the decreased chain flexibility of residues 112-118 of the CT molecule caused by the presence of cysteine at position 116. It is conceivable that the R116C mutation may have an effect on the motion of the semiflexible side chain containing R122 by interfering with the movement of oligopeptide chain into hydrolysis position and preventing an attack to the primary autolysis site at R122, resulting in an increased stability of trypsin[10]. Another potential mechanism is that the mutation results in disulfide-bridge formation with another proximal cysteine residue in the tertiary structure of trypsinogen polypeptide and, thus, plays a prominent role in increasing protein stability and decreasing its autolysis. However, none of these hypotheses has been critically studied.
The clinical manifestation of pancreatitis in this family merits some comments. Firstly, this is the first report of a family fulfilling the criteria for HP that has been shown to be due to R116C. The other previous three reports were mostly from sporadic symptomatic cases[10-12]. Our family confirms the dominant nature of this particular mutation. Secondly, this report exemplifies the intermittent nature of symptom shown in most family members rather than the chronic persistent symptom found in most HP. This phenomenon may not be common for all mutations and may be specific to this R116C mutation. This remains to be elucidated with further reports of similar and different mutations. Thirdly, the proband was later diagnosed with pancreatic cancer that was also suspected in her mother. This is contrary to the increased risk of pancreatic cancer found especially in the paternally inherited case[27]. If the diagnosis was certain, this might imply high cancer risk for this particular mutation that is different from the R122H mutation, which has negative contribution to the pathogenesis of a substantial fraction of pancreatic cancer[28]. It is well known that allelic heterogeneity of mutation may give rise to different clinical phenotypes. Cancer risks may therefore vary among different PRSS1 mutations. This may eventually lead to different choices of surgery depending upon the type of mutation and its associated cancer risk. Lastly, this family demonstrates variable age of onset, as well as incomplete penetrance for HP. These characteristics make diagnosis of HP in the first case difficult. From our experience with this family, there may be a benefit in justifying molecular testing in proband with very young age even though the history is not consistent with typical chronic pancreatitis.
ACKNOWLEDGEMENTS
We thank the family for their participation in this study. We also thank Visith Thongboonkerd for reading and commenting on the manuscript. Mahidol Research Grant to C.L had supported this work.
Science Editor Li WZ Language Editor Elsevier HK