Published online Jan 7, 2005. doi: 10.3748/wjg.v11.i1.84
Revised: February 28, 2004
Accepted: April 5, 2004
Published online: January 7, 2005
AIM: Transforming growth interacting factor (TGIF) is an inhibitor of both transforming growth factor β (TGF-β) and retinoid signaling pathways. Moreover, the activation of MAPK pathway can prolong its half-life. However, its role in carcinogenesis is still unknown. Thus we attempted to investigate the effect of TGIF on biologic behaviors of gastric carcinoma cells.
METHODS: Gastric carcinoma cell line, SGC-7901, was stably transfected with plasmid PcDNA3.1-TGIF. Western blotting and cell immunohistochemistry screening for the highly expressing clone of TGIF were employed. The growth of transfected cells was investigated by MTT and colony-formation assays, and apoptosis was measured by flow cytometry (FCM) and transmission electron microscopy. Tumorigenicity of the transfectant cells was also analyzed.
RESULTS: TGIF had no effect on the proliferation, cell cycle and apoptosis of SGC-7901 cells, but cellular organelles of cells transfected with TGIF were richer than those of vector control or parental cells. Its clones were smaller than the control ones in plate efficiency, and its tumor tissues also had no obvious necrosis compared with the vector control or parental cells. Moreover, TGIF could resist TGF-β mediated growth inhibition.
CONCLUSION: TGIF may induce differentiation of stomach neoplastic cells. In addition, TGIF can counteract the growth inhibition induced by TGF-β.
- Citation: Hu ZL, Wen JF, Xiao DS, Zhen H, Fu CY. Effects of transforming growth interacting factor on biological behaviors of gastric carcinoma cells. World J Gastroenterol 2005; 11(1): 84-88
- URL: https://www.wjgnet.com/1007-9327/full/v11/i1/84.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i1.84
Gastric carcinoma is one of the most frequent tumors that seriously threatens people’s health in China. However, its exact mechanism is still unclear. Many investigations have shown that most neoplasms are associated with TGF-β. Moreover, a variety of neoplasms are able to resist the growth inhibition mediated by TGF-β[1]. The role of TGF-β signaling pathway in the development of stomach neoplasms is worthy of our attention.
TGIF is a nucleoprotein that belongs to the homeobox domain TALE family, which has a three-amino acid insertion between helices 1 and 2 of the homeodomain[2]. TGIF locates on 18p11.3 and encodes a protein consisting of 272 amino acids. It has been discovered to be involved in many biological processes, such as human and Drosophila development. Recent studies have shown that Drosophila TGIF is essential for developmentally regulated transcription in its spermatogenesis[3,4]. Although TGIF homozygous mutant flies are viable and appear morphologically normal, the males are completely sterile. TGIF has also been identified as a small group of genes implicated in the human development disorder holoprosencephaly (HPE), a common structural defect of the developing forebrain in human being[5]. It has also been suggested to act as a competitive inhibitor of the TALE-class homeodomain protein Meis2 in neuronal cell lines[6].
In addition, TGIF also participates in a number of distinct pathways. Bertolino et al[2] discovered that TGIF could compete with retinoid for binding sites in promoter, and inhibit the retinoid signaling pathway, while retinoid could inhibit cell proliferation and induce cell differentiation and apoptosis. Recent studies reveal that TGIF is also a transcriptional co-repressor, which inhibits TGF-β signaling pathway[7-13].
TGIF inhibits TGF-β signaling pathway mainly by histone deacetylase (HDAC) dependent[7] and HDAC independent mechanisms[8]. In addition, TGIF also directly inhibits target gene expression via binding to DNA at its HD region consisting of 35-97 amino acid residues[10]. Recently Lo et al[11] revealed that TGF-β signaling pathway had a cross-talk with EGF/Ras/MAPK signaling pathway that could phosphorylate TGIF, prolong its half-life, and raise its protein level.
TGIF inhibits not only TGF-β signaling pathway but also retinoid signaling pathway. Moreover, EGF/Ras/MAPK signaling pathway prolong half-life of TGIF. But it is still unknown whether TGIF plays a role in carcinogenesis. To determine the role of TGIF in the carcinogenesis, SGC-7901, a moderately differentiated gastric carcinoma cell line, was transfected with TGIF to investigate the effect of TGIF on the biological behaviors of SGC-7901 cells.
The human gastric carcinoma cell line, SGC-7901, was cultured in RMPI 1640 medium containing 100 mL/L fetal bovine serum (FBS) supplemented with penicillin and streptomycin. Cultures were incubated in an incubator containing 5 mL/L CO2 in air at 37 °C.
Plasmid pcDNA3.1-TGIF was a gift from Professor Mouradian, Genetic Pharmacology Unit, Experimental Therapeutics Branch, NINDS, National Institutes of Health, Bethesda, Maryland 20892, USA.
Transfection and selection of positive clones were carried out in a 6-well plate. When the cells reached 70% confluence, the transfection process began. Briefly, solution A was prepared by diluting 2 μg of pcDNA3.1-TGIF into 200 μL serum-free medium, and solution B was prepared by diluting 5 μL lipofectamine (Life Technologies) into 200 μL serum-free medium. The two solutions were mixed for 20 min at room temperature, and then 0.6 mL serum-free medium was added to a tube containing the complex, and subsequently added to the rinsed cells. The medium was replaced with fresh and complete medium 18 h after transfection. Seventy-two hours after transfection, it was replaced again with the selective medium containing 800 g/L G418 (Alexis Biochemicals). Once stable transfection was obtained, the cells were maintained in 200 g/L of G418. Meanwhile, SGC-7901 cells were transfected with the empty pcDNA3.1 vector as control.
Total proteins were measured using the BCA kit (Pierce) according to the manufacturer’s protocol. Forty μg of total proteins was separated by 12% SDS-PAGE under reducing conditions, and transferred to nitrocellulose membrane. The nitrocellulose membrane was then incubated with blocking buffer (PBS containing 5% non-fat milk) for 2 h at room temperature and with goat polyclonal antibody against TGIF overnight at 37 °C with gentle shaking. The membrane was washed with PBS twice for 5 min, and then incubated with rabbit anti-goat IgG conjugated horseradish peroxidase diluted at 1:3000 (Zhongshan Co, Beijing) for 2 h at room temperature. After washing, TGIF was detected using DAB reagents. Ponceau S was used as a loading control.
Cells were seeded on slides at a density of 1.8×104 and grown for two days. Slides were washed once with PBS and fixed in acetone for 20 min at 4 °C. Fixed cells were washed 3 times with-PBS and nonspecific proteins were blocked using non-immune serum for 30 min at room temperature. Cells were incubated for 1 h with the goat polyclonal antibody against TGIF, then washed twice for 3 min with PBS. Cells were then incubated for 1 h with rabbit anti-goat IgG conjugated horseradish peroxidase diluted at 1:3000 (Zhongshan Co, Beijing), followed by two washes of 3 min in PBS. Cells were stained with DAB reagent.
Cells were cultured in 96-well plates at a density of 1×104 cells per well. Cell survival was measured by MTT assay 24, 48, 72, 96, 120 and 144 h after seeding. MTT assay was used to determine mitochondrial activity, which correlated with the number of viable cells in culture. Briefly, 20 μL of 5 g/L MTT (3-(4,5-dimethyl- thiazolyl-2)-2.5-diphenyltetrazolium bromide) in PBS was added to each well. Cells were incubated with MTT compound for 4 h at 37 °C in a 5 mL/L CO2 atmosphere, and subsequently 150 μL of DMSO was added to each well. The plates were incubated until MTT was completely resolved and A595 was measured.
Approximately 5×106 centrifugal sedimentation cells were immediately fixed in 700 mL/L ethanol and stored at 4 °C in PBS for fluorescence-activated cell sorting. Flow cytometry analysis was performed on a FACStar flow cytometer (Becton Dickinson). Histograms of cell number logarithmic fluorescence intensity were recorded for 10000 cells per sample.
To determine plating efficiency, cells were seeded in 6-well plates, 1000 cells per well. After 14 d, the colonies were fixed with 4% methanol and stained with 5% Giemsa solution (Sigma). The number of colonies with a diameter larger than 1 mm was counted. The plating efficiency (PE) was calculated as follows: PE = (colonies formed/cells seeded) ×100%.
Pellet of the transfected cells was fixed in 2.5 g/L glutaraldehyde, postfixed with 10 g/L osmium tetroxide, treated with 20 g/L uranyl acetate, dehydrated in ethanol, infiltrated with propylene oxide, and embedded in Epon mixture. Ultrathin sections were observed under Opton EM 10C (Germany).
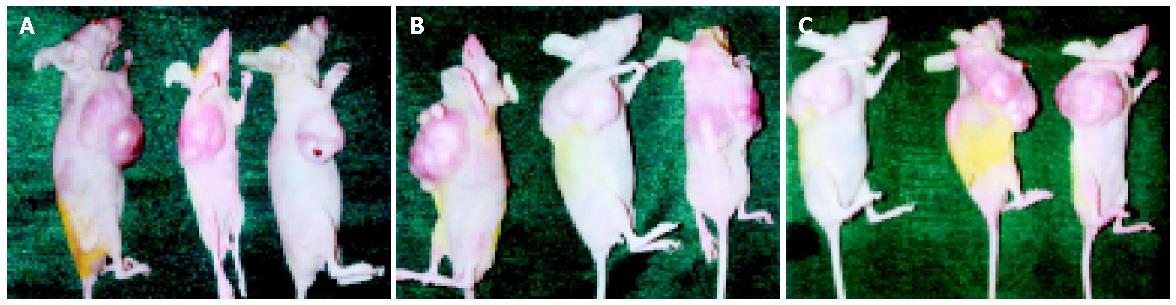
Nine female nude mice (BALB/c-nude, 4-6 wk old, weighing 16-18 g) were divided into 3 groups, 3 mice each group, and inoculated subcutaneously at the left flank with TGIF transfectant, vector control and parental cells (7×106 cells suspended in 0.2 mL of phosphate-buffered saline) and monitored for tumor development. Tumor size and animal weight were measured weekly. The nude mice were sacrificed and tumors were removed 35 d after inoculation.
F test was used. P value less than 0.05 was considered statistically significant.
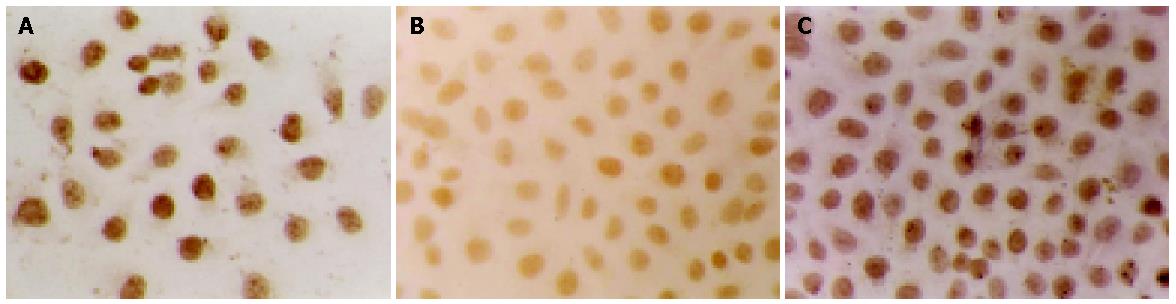
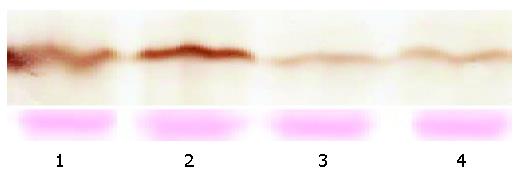
After transfection of SGC-7901 cells with a vector encoding TGIF, we identified two cell clones that constitutively overexpressed this protein by cell immunohistochemistry (Figure 1) and Western blot analyses (Figure 2). We selected one of them for further experiment.
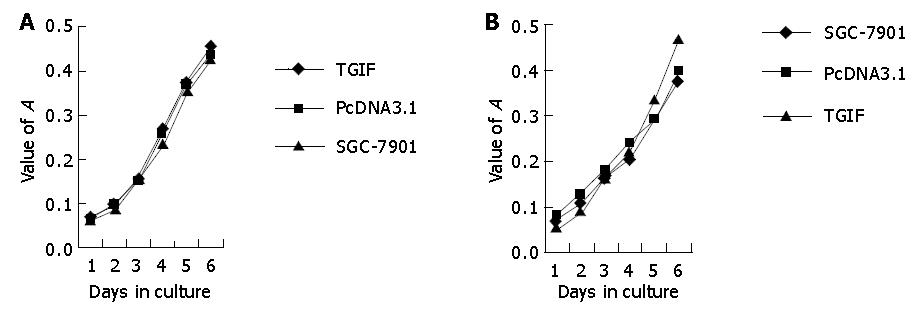
To determine the impact of TGIF on cell growth in vitro, we examined the rate of cell growth with MTT assay. As shown in Figure 3A, the growth rate of cells overexpressing TGIF had no distinct changes compared with blank and negative controls. After incubating with 10 ng/mL TGF-β 1 for the indicated time, both parental and vector control cells were found to have lower growth rates. However, TGIF transfected cells had no distinct changes (Figure 3B) and the difference was significant (P<0.05).
To confirm the effect of TGIF on proliferation of SGC-7901 cells, the cell cycle distribution and apoptosis were determined by flow cytometry. As shown in Table 1, TGIF had no effect on cell cycle and apoptosis rate of SGC-7901 cells. In parental and vector control cells, cell content of G1 phase obviously increased after treatment with TGF-β 1. However, TGIF expressing cells had no distinct change in cell content of G1 phase. All groups had a slight increase in apoptosis rate after incubation with 10 μg/L TGF-β 1 for 72 h.
| TGF-β 1 (-) (%) | TGF-β 1 (+) (%) | |||||||
| G1 | G2 | S | Apoptosis | G1 | G2 | S | Apoptosis | |
| SGC-7901 | 55.2 | 15.8 | 29 | 1.35 | 63.2 | 14.2 | 22.6 | 2.7 |
| PcDNA3.1 | 50.1 | 15.8 | 34.2 | 0.36 | 60.3 | 13.9 | 25.8 | 0.54 |
| TGIF | 50.9 | 18.4 | 30.6 | 0.41 | 50.1 | 21.2 | 28.7 | 0.67 |
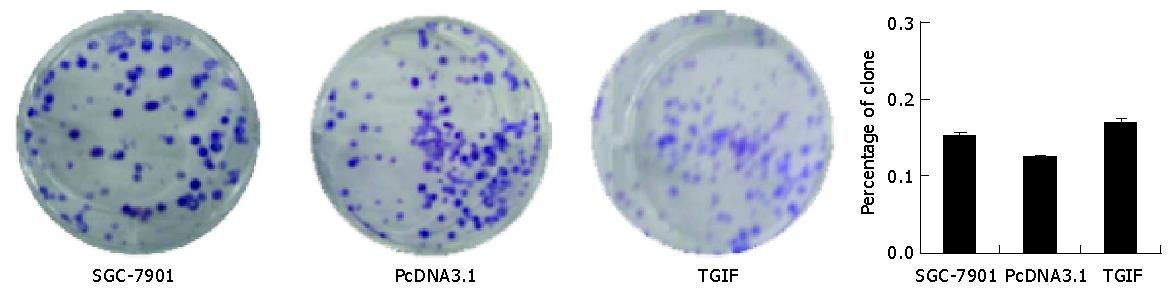
Plating efficiency in parent, vector control and TGIF transfected cells was 15.1%, 12.4% and 16.9% respectively (Figure 4), and the difference had no statistical significance (P>0.05). However, clones of TGIF transfected cells were smaller than those of the parental and vector control cells (Figure 4, upper panel).
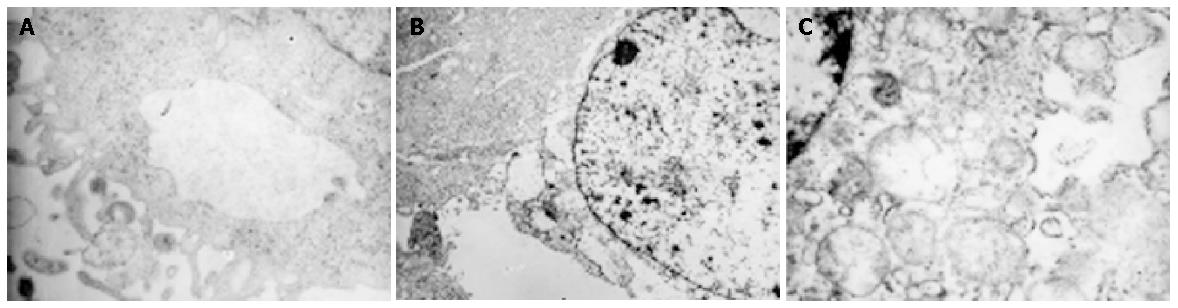
Apoptotic body was not found in parental, vector control or TGIF transfected cells under TEM, but there were more cell organellae in TGIF transfected cells compared with the blank and negative control cells (Figure 5).
We examined the effect of TGIF expression on SGC-7901 cell growth in athymic mice. Tumors were palpable in the first week after inoculation of cells in female athymic mice. As shown in Figure 6, TGIF transfectants revealed no difference in tumor growth as compared with vector control and parental cells throughout the observation period. The mean tumor weights in mice transfected parental, vector control cells and TGIF were 0.85±0.09, 0.87±0.13 and 0.87±0.27 g, respectively. In addition, the mean tumor volumes were 0.99±0.08, 1.01±0.11 and 1.10±0.12 cm3, respectively. However, the differences had no statistical significance (P>0.05). The animals were killed in accordance with the institutional tumor burden guidelines. After tumors were excised, there were necrotic tissues effused from tumors in parental and vector control groups but not in TGIF transfectants.
TGIF is a transcription factor that has been implicated in a number of distinct pathways. TGIF was first identified as a competitor of retinoic acid receptor to bind to retinoic acid response elements[2]. Subsequently TGIF interacts with Smads and is an inhibitor of TGF-β signaling pathway[7-13]. Recently Lo et al[11] revealed that MAPK signaling pathway had a cross-talk with TGF-β signaling pathway. MAPK transducing pathway can phosphorylate TGIF, prolong its half-life and raise its protein level. The enhancement of TGIF function might inhibit negative regulation of cell cycle by TGF-β. Its role in tumorigenicity is worthy of attention. Several reports have indicated that TGIF probably implicates in carcinogenesis. Nakakuki et al[14] discovered that TGIF gene was overexpressed in esophageal carcinoma. Voorter et al[15] revealed that there was gene amplification at 18p11 where TGIF locates in bladder transitional cell carcinoma using comparative genome hybridization. Luo et al[16] found that autologous antibody against TGIF existed in serum of patients with ovarian carcinoma.
Our experiment revealed that the growth rate of SGC-7901 cells had no distinct difference after transfection with TGIF (Figure 3), and the distribution of cell cycle had no obvious change either (Table 1). This result is not consistent with Edwards’ report showing that overexpression of TGIF overcame the checkpoint of yeast G1 phase[17]. The distinction may attribute to the difference of cell types. There were no differences in plating efficiency and nude mice tumorigenicity among parental, vector control and TGIF transfected cells (Figures 4 and 6). All these indicate that TGIF cannot worsen the biological behavior of SGC-7901 cells. Conversely, the number of cell organelles in TGIF transfected cells increased compared to blank and negative control cells (Figure 5), and the tumor tissues of TGIF transfectant group exhibited no distinct necrosis compared to control groups. This data indicates that TGIF may induce differentiation of SGC-7901 cells, at least in part.
After the treatment with TGF-β 1, parental and vector control cells showed distinct reduction in cell growth, whereas TGIF transfectants revealed no obvious difference (Figure 3). Flow cytometry also showed similar results (Table 1). Our finding is coincident with Lo’s report that HaCaT cell line could resist the growth inhibition mediated by TGF-β after stable transfection of TGIF[11]. Thus overexpression of TGIF protein can inhibit the negative regulation of TGF-β in cell cycle. It also implies that tumor cells may escape the growth inhibition by TGF-β via this mechanism.
Cells transfected with TGIF showed no distinct difference in apoptosis compared to the controls (Table 1). After incubation with TGF-β 1 for 72 h, there was no distinct difference among the three groups, indicating that TGIF may not interfere with TGF-β-mediated cell apoptosis. However, we cannot rule out the possibility that the insensitivity to TGF-β-mediated apoptosis resulting from the disturbance of TGF-β signaling pathway in SGC-7901 cells, contributes to cells transfected with TGIF resisting TGF-β mediated apoptosis.
Edited by Ma JY and Wang XL Proofread by Zhu LH
| 1. | Wieser R. The transforming growth factor-beta signaling pathway in tumorigenesis. Curr Opin Oncol. 2001;13:70-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 2. | Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178-31188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 222] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 3. | Wang Z, Mann RS. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development. 2003;130:2853-2865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Ayyar S, Jiang J, Collu A, White-Cooper H, White RA. Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development. 2003;130:2841-2852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massagué J, Muenke M. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25:205-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 284] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 6. | Yang Y, Hwang CK, D'Souza UM, Lee SH, Junn E, Mouradian MM. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J Biol Chem. 2000;275:20734-20741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Wotton D, Lo RS, Lee S, Massagué J. A Smad transcriptional corepressor. Cell. 1999;97:29-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 436] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 8. | Melhuish TA, Wotton D. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem. 2000;275:39762-39766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 84] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Wotton D, Knoepfler PS, Laherty CD, Eisenman RN, Massagué J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12:457-463. [PubMed] |
| 10. | Wotton D, Lo RS, Swaby LA, Massagué J. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J Biol Chem. 1999;274:37105-37110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 145] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 11. | Lo RS, Wotton D, Massagué J. Epidermal growth factor signaling via Ras controls the Smad transcriptional co-repressor TGIF. EMBO J. 2001;20:128-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 129] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Pessah M, Prunier C, Marais J, Ferrand N, Mazars A, Lallemand F, Gauthier JM, Atfi A. c-Jun interacts with the corepressor TG-interacting factor (TGIF) to suppress Smad2 transcriptional activity. Proc Natl Acad Sci USA. 2001;98:6198-6203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Chen F, Ogawa K, Nagarajan RP, Zhang M, Kuang C, Chen Y. Regulation of TG-interacting factor by transforming growth factor-beta. Biochem J. 2003;371:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Nakakuki K, Imoto I, Pimkhaokham A, Fukuda Y, Shimada Y, Imamura M, Amagasa T, Inazawa J. Novel targets for the 18p11.3 amplification frequently observed in esophageal squamous cell carcinomas. Carcinogenesis. 2002;23:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Voorter C, Joos S, Bringuier PP, Vallinga M, Poddighe P, Schalken J, du Manoir S, Ramaekers F, Lichter P, Hopman A. Detection of chromosomal imbalances in transitional cell carcinoma of the bladder by comparative genomic hybridization. Am J Pathol. 1995;146:1341-1354. [PubMed] |
| 16. | Luo LY, Herrera I, Soosaipillai A, Diamandis EP. Identification of heat shock protein 90 and other proteins as tumour antigens by serological screening of an ovarian carcinoma expression library. Br J Cancer. 2002;87:339-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |