Published online Jan 7, 2005. doi: 10.3748/wjg.v11.i1.31
Revised: April 21, 2004
Accepted: May 13, 2004
Published online: January 7, 2005
AIM: To detect the variations of mitochondrial 12S rRNA in patients with gastric carcinoma, and to study their significance and the relationship between these variations and the genesis of gastric carcinoma.
METHODS: PCR amplified mitochondrial 12S rRNA of 44 samples including 22 from gastric carcinoma tissues and 22 from adjacent normal tissues, was detected by direct DNA sequencing. Then laser capture microdissection technique (LCM) was used to separate the cancerous cells and dysplasia cells with specific mutations. Denaturing high performance liquid chromatography (DHPLC) plus allele-specific PCR (AS-PCR), nest-PCR and polyacrylamide gel electrophoresis (PAGE) were used to further evaluate this mutant property and quantitative difference of mutant type between cancerous and dysplasia cells. Finally, RNAdraw biosoft was used to analyze the RNA secondary structure of mutant-type 12S rRNA.
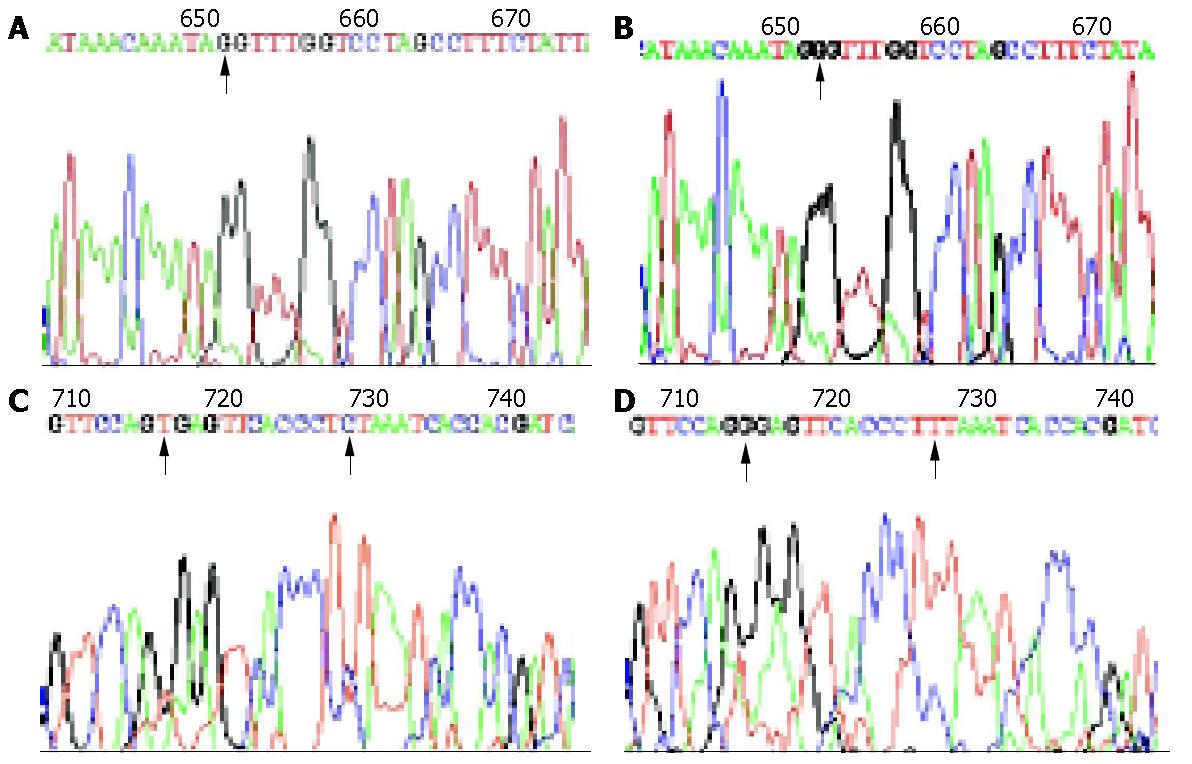
RESULTS: Compared with Mitomap database, some new variations were found, among which np652 G insertion and np716 T-G transversion were found only in cancerous tissues. There was a statistic difference in the frequency of 12S rRNA variation between intestinal type (12/17, 70.59%) and diffusive type (5/17, 29.41%) of gastric carcinoma (P<0.05). DHPLC analysis showed that 12S rRNA np652 G insertion and np716 T-G transversion were heteroplasmic mutations. The frequency of 12S rRNA variation in cancerous cells was higher than that in dysplasia cells (P<0.01). 12S rRNA np652 G insertion showed obviously negative effects on the stability of 12S rRNA secondary structure, while others such as T-G transversion did not.
CONCLUSION: The mutations of mitochondrial 12S rRNA may be associated with the occurrence of intestinal-type gastric carcinoma. Most variations exist both in gastric carcinomas and in normal tissues, and they might not be the characteristics of tumors. However, np652 G insertion and np716 T-G transversion may possess some molecular significance in gastric carcinogenesis. During the process from normality to dysplasia, then to carcinoma, 12S rRNA tends to convert from homoplasmy (wild type) to heteroplasmy, then to homoplasmy (mutant type, np717 T-G).
- Citation: Han CB, Ma JM, Xin Y, Mao XY, Zhao YJ, Wu DY, Zhang SM, Zhang YK. Mutations of mitochondrial 12S rRNA in gastric carcinoma and their significance. World J Gastroenterol 2005; 11(1): 31-35
- URL: https://www.wjgnet.com/1007-9327/full/v11/i1/31.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i1.31
It is known that gastric carcinogenesis is associated with some oncogenes and tumor suppressor genes, but no definite mechanism is understood. Mitochondria not only are the place where biological oxidation occurs, but also participate in cell apoptosis[1,2]. In recent researches, mitochondrial DNA (mtDNA) has been considered to be associated with tumor-genesis[3-8]. The mtDNA is a 16569-bp double-stranded, closed circular molecule, which encodes polypeptides participating in oxidative phosphorylation and synthesis of ATP. However, unlike nuclear DNA, mtDNA is more susceptible to damage by exogenous mutagens and endogenous factors owing to many reasons, such as lack of protection by histones and an effective mismatch repair (MMR) system[9,10], and the internal environment with high levels of free radicals and reactive oxygen species (ROS) generated from oxygen in the organelle[11,12]. 12S rRNA is one of the two riboproteosome genes (12S rRNA and 16S rRNA) in mtDNA, with conserved sequences and no introns. 12S rRNA transcripts, together with the other ribosome proteins encoded by nuclear genes, are composed of the small subunits of mitochondrial ribosome[13]. Once there are mutations occurring in 12S rRNA, it might result in abnormal assembly of small subunits and affect the synthesis of mitochondrial proteins such as cyt-c, and even influence cell apoptosis. Thus we carried out an in-depth study on the mutations in 12S rRNA and their significance in gastric carcinogenesis. Multiple techniques were utilized in our study including DNA sequencing, laser capture microdissection (LCM), allele specific PCR (AS-PCR), nest-PCR and denaturing high performance liquid chromatography (DHPLC) to help us get more confirmed results.
A total of 44 gastric carcinoma tissues and adjacent normal gastric mucosal tissues were obtained from the specimens of 22 patients with gastric carcinoma in the First Affiliated Hospital of China Medical University. All tissues were immediately frozen in liquid nitrogen after resection, stored at -80 °C, and diagnosed according to WHO’s histological classification and Lauren’s classification of gastric carcinoma. Twelve samples were intestinal type and the other ten were diffusive type.
nDNA and mtDNA extraction Thirty micrograms of gastric epithelial mucosal tissue was homogenized in a homogenizer for 30 s, and then digested with 1 mL of extraction buffer (0.1 g/L proteinase K, 10 mmol/L Tris-HCl, 0.1 mol/L EDTA pH 7.4, 5 g/L SDS). DNA was extracted with phenol/chloroform/ isopropanol and precipitated in ethanol. The precipitated DNA was recovered in 50 μL of 10 mmol/L Tris-HCl, 0.1 mmol/L EDTA (pH 8.0). Each DNA sample was balanced in concentration using a spectrophotometer.
PCR amplification of 12S rRNA PCR amplification was carried out in a final volume of 50 μL, containing 50 ng DNA, 0.5 μmol/L of each primer P1 (Table 1) 2.5 mmol/L MgCl2, 200 μmol/L of each dNTP, and 2.5U Taq DNA polymerase (TaKaRa Ex TaqTM). Amplification was performed in a Biometra personal PCR system. The amplification conditions were as follows: an initial incubation at 94 °C for 4 min, followed by 30 cycles, each consisting of denaturing at 94 °C for 45 s, annealing at 60 °C for 45 s and extension at 72 °C for 1 min, and a final step of extension at 72 °C for 3 min.
| Primer | Forward primer | Position | Reverse primer |
| P1 | TAGACGGGCTCACATCAC | 620 | GGGGTATCTAATAGTTTGGGT |
| P2 | GCAAGCATCCCCGTTCCAGT | 697 | GGGGTATCTAATAGTTTGGGT |
| P3 | GCAAGCATCCCCGTTCCAGG | 697 | GGGGTATCTAATAGTTTGGGT |
DNA sequencing PCR products were sent to the United Gene Technology Company Limited, Shanghai, China, for direct sequencing (3 700, Bigdye-Terminator), and the results were compared with PubMed database and Mitomap mitochondrial database.
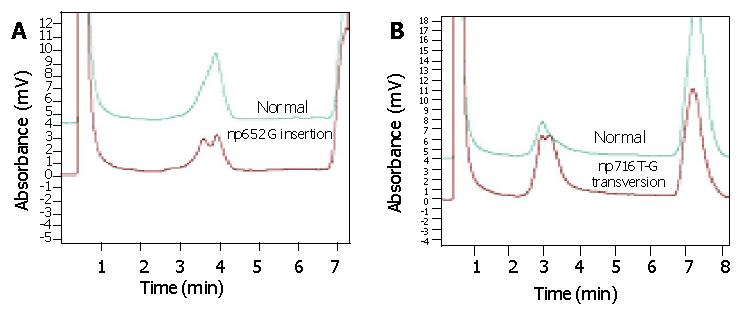
Denaturing high performance liquid chromatography Compared with nuclear genome, the mutation of mtDNA had more heteroplasmy. Therefore, denaturing high performance liquid chromatography (DHPLC) was performed to detect the property of the specific mutation 716 T-G found in 12S rRNA, in order to analyze whether the mutation was homoplasmic mutation (pure mutant) or heteroplasmic mutation (wild and mutant type). The PCR products were denatured at 95 °C for 5 min, then annealed gradually to 65 °C at the speed of 0.1 °C/s. The annealed samples were placed on the 96-well plate of WAVETM chromatography instrument (Transgenomic, America). The appropriate reaction temperature was calculated via Wave Maker 4.1 software (America) according to the imported DNA sequences, and then the samples were imported at the speed of 0.9 mL/min. Double peaks were regarded as heteroplasmic variation, while a single peak as homoplasmic variation.
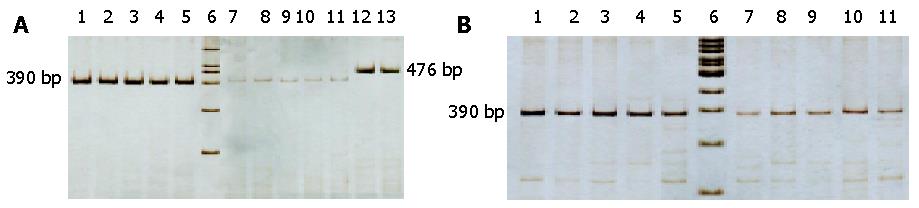
Nest-PCR and AS-PCR Nest-PCR and allele-specific PCR were performed to demonstrate whether there were some quantitative differences in mutant 12S rRNA between gastric cancerous cells and dysplasia cells, which were diagnosed pathologically and microdissected by LCM (PixCell II LCM 200, America). Primers, P2 and P3, were designed according to 12S rRNA 716T-G mutations (Table 1). P1 served as the outer primer of nest-PCR, and was used to amplify 12S rRNA from the samples of wild type and mutant type in gastric cancerous and dysplasia cells respectively. After purified by gel electrophoresis and quantified, these PCR products were diluted to 10 mg/L. Five microliters of the dilution were taken as the template for the second reaction. We amplified the wild type (labeled as PP2) and mutant type with 716T-G mutation (labeled as PP3) with inner primers P2 and P3. Every PCR reaction was repeated five times, then the mean value was calculated. T3 and D3, values of PP3/PP3+PP2 in carcinoma and dysplasia respectively, were calculated and statistically compared to analyze the quantitative difference in mutant 12S rRNA with 716T-G mutation between cancerous cells and dysplasia cells. Conditions of PCR reaction were the same as above. Reaction system and condition were rigidly kept consistent.
PAGE and silver staining Polyacrylamide gel electrophoresis (PAGE) was performed with 3% concentrated gel and 8% separated gel. Five microliters of mixture, containing 3 μL PCR product and 2 μL sampling buffer, were run for electrophoresis at 8 mA for 20 min, and then at 15 mA for 30 min. Silver staining procedure was in reference to literature[14,15]. Gel was scanned with the ChemiImager and quantified with Image J software (Sweden).
RNA secondary structure analysis RNAdraw 1.1b2 (USA) was used to predict the RNA secondary structures of mutant type and wild type of 12S rRNA(T = 37 °C).
All data were processed by SPSS version 10.0. Quantitative difference and variant frequencies between different groups were analyzed with t-test and chi-square test respectively. P<0.05 was considered statistically significant. Quantitative data are expressed as mean±SD.
After searching the PubMed and Mitomap, we found that there also were some variations in conserved 12S rRNA regions (Table 2). Some of the variations were first reported (Figure 1), among which np652 G insertion and np716 T-G transversion were found only in cancerous tissues. It showed a higher variation frequency in intestinal type of gastric carcinoma (12/17, 70.59%) than in diffuse type of gastric carcinoma (5/17, 29.41%) (P = 0.019). But the difference in variation frequency between normal gastric tissues in (15/22) and cancerous tissues (17/22) had no statistical significance (P>0.05).
| Position (np) | Variations | Property |
| 652 | G-ins1 | HM |
| 663 | A-G | PM |
| 709 | G-A | PM |
| 716 | T-G1 | HM |
| 728 | C-T | PM |
| 745 | A-G1 | PM |
| 750 | G-A | PM |
| 757 | A-T | PM |
| 759 | A-T1 | PM |
| 764 | A-C | PM |
| 772 | A-T1 | PM |
| 782 | A-T1 | PM |
| 794 | T-C1 | PM |
| 799 | A-C1 | PM |
DHPLC verified that np652G insertion and np716T-G transversion mutation were heteroplasmic mutations mixed with mutant type and wild type 12S rRNA (Figure 2). AS-PCR, nest-PCR, PAGE and silver staining further showed that 12S rRNA with 716 T-G mutation was predominantly accompanied with a small number of wild type 12S rRNAs without 716T-G mutation (Figure 3). Repeated AS-PCR showed that there was a statistic difference in mutant 12S rRNA between gastric cancerous cells and dysplasia cells, T3 = 0.89±0.02 (Table 3) and D3 = 0.68±0.09 (P<0.01). Quantitative analysis of np652 G insertion was not performed because it was not suitable for AS-PCR and had no corresponding endonuclease.
| Num. | Cancer (T3) | Dysplasia (D3) |
| 1 | 0.9214 | 0.8164 |
| 2 | 0.8738 | 0.6487 |
| 3 | 0.9038 | 0.7268 |
| 4 | 0.8955 | 0.5572 |
| 5 | 0.8659 | 0.6402 |
| Mean±D | 0.89±0.02 | 0.68±0.09 |
| P | <0.01 |
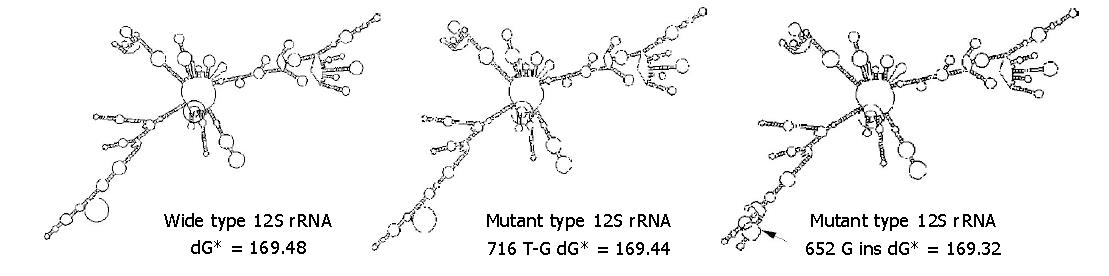
The 652G insertion significantly influenced 12S rRNA secondary structure, while 716 T-G transversion and other variations had only limited effects (Figure 4). The secondary structure free energy (dG) of mutant type with 716 T-G, 652 G insertions and wild type was 169.44 kcal/mol (dG-mL), 169.32 kcal/mol (dG-m2) and 169.48 kcal/mol (dG-w), respectively.ΔdG between wild type and mutant type (subtractions of dG-w and dG-mL, dG-m2 respectively) was 0.04 kcal/mol and 0.16 kcal/mol. Considering the correlation between energy and structural stability, np652 G insertion was more disadvantageous to the stability of 12S rRNA RNA secondary structure.
The existing research data indicates that the biological features of tumors are not only decided by nuclear genetic materials, but also related with extranuclear mtDNA[5-10,16-19]. The mtDNA could not protect histones, and synthesize glutathione to eliminate the oxygen free radicals generated by oxidative phosphorylation[18]. Therefore, mtDNA is susceptible to the attack by harmful endogenous factors and exogenous carcinogens. Lipidophilic carcinogens preferentially aggregate at mtDNA because of the high ratio of lipid and DNA in mitochondria. In addition, the limited proofreading capacity of mtDNA polymerase γ and hairpin structure in tRNA genes make more replicative mismatches during the process of mtDNA replication than that in nuclear DNA[19]. Nowadays, mitochondrial mismatch repair (MMR) system has been found only in yeasts, MSH1 is involved in mitochondrial genome repair system, and MSH2 in nuclear DNA repair system. But no MSH1 homologue has been found in mammalian cells, and therefore it is widely accepted that mitochondrion lacks MMR system[20]. Since mtDNA is the major target attacked by carcinogens, the injury severity and mutant rate of mtDNA are significantly higher than those of nuclear DNA (around 10 times)[21-24].
The study showed that most of the 12S rRNA variations in gastric carcinomas were not tumor-specific, but they might be one of the latent risk genetic changes for gastric carcinogenesis. Endogenous and exogenous damage factors might lead to mitochondrial oxygen stress and increase ROS content under certain conditions, which could damage mtDNA and mitochondrial lipid membrane structure, impede protein synthesis, electron transport and ATP synthesis, and even influence cyt-c release channel, voltage-dependent anion channel in the mitochondrial outer membrane, and cell apoptosis. Intestinal type of gastric carcinogenesis is closely associated with environmental factors[25-27]. The intestinal type of gastric carcinoma is predominant in high incidence regions, which has an important epidemiological significance. While diffusive type is more related with heredity[28,29]. Mutant frequencies of 12S rRNA existing in intestinal type of gastric carcinoma are higher than those in diffusive type (P<0.05), indicating that a higher variation of mtDNA might play an important role in intestinal-type gastric carcinogenesis, consistent with the fact that mitochondria and mtDNA are susceptible to the harmful environmental factors. Habano et al[30,31] discovered that 10/62 (16%) cases of gastric cancers had the polyC instability in mitochondrial control region(D-loop), 5 cases had point mutations in patients with polyC instability. Mitochondrial genomic instability (mtGI) frequently occurs in intestinal type gastric carcinoma. Like nuclear MSI, mitochondrial variations might also be related with mtGI.
12S rRNA np716T-G transversion and np652G insertion are two specific mutations found in tumor mtDNA. The np716 T-G transversion is a heteroplasmic mutation. During the process of normality to dysplasia, then to carcinoma, 12S rRNA tends to convert from homoplasmy (wild type) to heteroplasmy, then to homoplasmy (mutant type, np717 T-G), suggesting that during the development from precancerous lesion to cancer, some mutations in mtDNA aggregate in descent cells, which participate in gastric carcinogenesis. The aggregated mutant mtDNA might be the sequent result that fewer single copies of mutant mtDNA are produced by damages and increase in quantity after several cell divisions. The wild type and mutant type of mtDNA tend to form their aggregating domains during cell division, thereby it is possible that two descent cells inherit wild type and mutant type of mtDNA respectively. Furthermore, mutant mtDNA might bear a replicative superiority[32], the cells containing mutant mtDNA would be predominant in the whole cell group (homoplasmic mutant type) because of the clone growth superiority.
Unlike the translation initiation of mRNA in cytoplasm, mtRNA in mitochondria has no up stream promoter sequence which binds to ribosomal small subunits, the small subunits have to bind beside the N-formylmethionine at 5' end. Ribosomal 28S small subunits need 30-80 bp nucleotides to interact with mtRNA, and at least 400 bp to effectively bind to each other, which could lead to decreased translation efficiency[13]. np562 G insertion could lead to the change of 12S rRNA secondary structure, which might influence the normal function of small subunits, and induce the lower translation efficiency. Because all the 13 proteins encoded by mitochondrial DNA participate in oxidative phosphorylation and ATP synthesis, low translation efficiency surely reduces synthesis of these proteins, thus causing subsequent cell energy loss, change of membrane permeability and a series of molecular changes.
In conclusion, intestinal gastric carcinoma is related to the variations of mitochondrial 12S rRNA gene. Though most of the variations are not tumor-specific, they might be a latent risk factor for gastric carcinogenesis because impairment of mtDNA is correlated with environmental and biological factors. Therefore, our results provide a new pathway for the research on molecular mechanisms of intestinal gastric carcinogenesis.
Edited by Wang XL and Zhu LH
| 1. | Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3067] [Cited by in RCA: 2970] [Article Influence: 114.2] [Reference Citation Analysis (0)] |
| 2. | Máximo V, Soares P, Seruca R, Sobrinho-Simões M. Comments on: mutations in mitochondrial control region DNA in gastric tumours of Japanese patients, Tamura, et al. Eur J Cancer 1999, 35, 316-319. Eur J Cancer. 1999;35:1407-1408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Bianchi NO, Bianchi MS, Richard SM. Mitochondrial genome instability in human cancers. Mutat Res. 2001;488:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 131] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Máximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simões M. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer. 2001;32:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 5. | Bianchi MS, Bianchi NO, Bailliet G. Mitochondrial DNA mutations in normal and tumor tissues from breast cancer patients. Cytogenet Cell Genet. 1995;71:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Asumendi A, Morales MC, Alvarez A, Aréchaga J, Pérez-Yarza G. Implication of mitochondria-derived ROS and cardiolipin peroxidation in N-(4-hydroxyphenyl)retinamide-induced apoptosis. Br J Cancer. 2002;86:1951-1956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 7. | Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 618] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 8. | Nishikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 2001;61:1843-1845. [PubMed] |
| 9. | Kotake K, Nonami T, Kurokawa T, Nakao A, Murakami T, Shimomura Y. Human livers with cirrhosis and hepatocellular carcinoma have less mitochondrial DNA deletion than normal human livers. Life Sci. 1999;64:1785-1791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, Guzman A, Oliva H, Sancho M. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 11. | Lee I, Bender E, Kadenbach B. Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol Cell Biochem. 2002;234-235:63-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 113] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Atlante A, Calissano P, Bobba A, Azzariti A, Marra E, Passarella S. Cytochrome c is released from mitochondria in a reactive oxygen species (ROS)-dependent fashion and can operate as a ROS scavenger and as a respiratory substrate in cerebellar neurons undergoing excitotoxic death. J Biol Chem. 2000;275:37159-37166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 159] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 995] [Cited by in RCA: 1063] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 14. | Han CB, Zhao YJ, Li F, He Q, Ma JM, Xin Y. Quantitation and detection of deletion in tumor mitochondrial DNA by microarray technique. Zhonghua ZhongLiu ZaZhi. 2004;26:10-13. [PubMed] |
| 15. | Han CB, Li F, Zhao YJ, Ma JM, Wu DY, Zhang YK, Xin Y. Variations of mitochondrial D-loop region plus downstream gene 1 2S rRNA-tRNA(phe) and gastric carcinomas. World J Gastroenterol. 2003;9:1925-1929. [PubMed] |
| 16. | Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. Detection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patients. Int J Cancer. 2001;94:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Tamura G, Nishizuka S, Maesawa C, Suzuki Y, Iwaya T, Sakata K, Endoh Y, Motoyama T. Mutations in mitochondrial control region DNA in gastric tumours of Japanese patients. Eur J Cancer. 1999;35:316-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Herrera B, Alvarez AM, Sánchez A, Fernández M, Roncero C, Benito M, Fabregat I. Reactive oxygen species (ROS) mediates the mitochondrial-dependent apoptosis induced by transforming growth factor (beta) in fetal hepatocytes. FASEB J. 2001;15:741-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 241] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 19. | Pinz KG, Shibutani S, Bogenhagen DF. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J Biol Chem. 1995;270:9202-9206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 89] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975-985. [PubMed] |
| 21. | Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482-1488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2207] [Cited by in RCA: 2189] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 22. | Haga N, Fujita N, Tsuruo T. Mitochondrial aggregation precedes cytochrome c release from mitochondria during apoptosis. Oncogene. 2003;22:5579-5585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Heddi A, Stepien G, Benke PJ, Wallace DC. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J Biol Chem. 1999;274:22968-22976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 131] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Chung YM, Bae YS, Lee SY. Molecular ordering of ROS production, mitochondrial changes, and caspase activation during sodium salicylate-induced apoptosis. Free Radic Biol Med. 2003;34:434-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 25. | Flucke U, Mönig SP, Baldus SE, Zirbes TK, Bollschweiler E, Thiele J, Dienes HP, Hölscher AH. Differences between biopsy- or specimen-related Laurén and World Health Organization classification in gastric cancer. World J Surg. 2002;26:137-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Zirbes TK, Baldus SE, Moenig SP, Schmitz K, Thiele J, Holscher AH, Dienes HP. Tenascin expression in gastric cancer with special emphasis on the WHO-, Lauren-, and Goseki-classifications. Int J Mol Med. 1999;4:39-42. [PubMed] |
| 27. | Pinheiro PS, van der Heijden LH, Coebergh JW. Unchanged survival of gastric cancer in the southeastern Netherlands since 1982: result of differential trends in incidence according to Laurén type and subsite. Int J Cancer. 1999;84:28-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 28. | Zhou HP, Wang X, Zhang NZ. Early apoptosis in intestinal and diffuse gastric carcinomas. World J Gastroenterol. 2000;6:898-901. [PubMed] |
| 29. | Lee KE, Lee HJ, Kim YH, Yu HJ, Yang HK, Kim WH, Lee KU, Choe KJ, Kim JP. Prognostic significance of p53, nm23, PCNA and c-erbB-2 in gastric cancer. Jpn J Clin Oncol. 2003;33:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Habano W, Nakamura S, Sugai T. Microsatellite instability in the mitochondrial DNA of colorectal carcinomas: evidence for mismatch repair systems in mitochondrial genome. Oncogene. 1998;17:1931-1937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 31. | Habano W, Sugai T, Nakamura SI, Uesugi N, Yoshida T, Sasou S. Microsatellite instability and mutation of mitochondrial and nuclear DNA in gastric carcinoma. Gastroenterology. 2000;118:835-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 96] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | Penta JS, Johnson FM, Wachsman JT, Copeland WC. Mitochondrial DNA in human malignancy. Mutat Res. 2001;488:119-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 325] [Article Influence: 13.5] [Reference Citation Analysis (0)] |