Published online Jan 7, 2005. doi: 10.3748/wjg.v11.i1.118
Revised: April 8, 2004
Accepted: May 13, 2004
Published online: January 7, 2005
AIM: To obtain the active human recombinant uridine diphosphate glucuronosyltransferase 1A3 (UGT1A3) enzyme from Chinese hamster lung (CHL) cells.
METHODS: The full-length UGT1A3 gene was amplified by reverse transcription-polymerase chain reaction (RT-PCR) using total RNA from human liver as template. The correct fragment confirmed by sequencing was subcloned into the mammalian expression vector pcDNA3.1 (+), and the recombinant vector was transfected into CHL cells using a calcium phosphate method. Expressed UGT1A3 protein was prepared from CHL cells resistant to neomycin (G418). Then the protein was added into a reaction mixture for glucuronidation of quercetin. The glucuronidation activity of UGT1A3 was determined by reverse phase-high performance liquid chromatography (RP-HPLC) coupled with a diode array detector (DAD). The quercetin glucuronide was confirmed by hydrolysis with β-glucuronidase. Control experiments were performed in parallel. The transcriptions of recombinants were also determined by RT-PCR.
RESULTS: The gene was confirmed to be an allele (UGT1A3-3) of UGT1A3 by DNA sequencing. The fragment was introduced into pcDNA3.1 (+) successfully. Several colonies were obtained under the selection pressure of G418. The result of RT-PCR showed transcription of recombinants in mRNA level. Glucuronidation assay and HPLC analysis indicated UGT1A3 expressed heterologously in CHL cells was in an active form, and one of the gulcuronides corresponding to quercetin was also detected.
CONCLUSION: Correct sequence of UGT1A3 gene can be obtained, and active UGT1A3 enzyme is expressed heterologously in CHL cells.
- Citation: Chen YK, Li X, Chen SQ, Zeng S. Heterologous expression of active human uridine diphosphate glucuronosyltransferase 1A3 in Chinese hamster lung cells. World J Gastroenterol 2005; 11(1): 118-121
- URL: https://www.wjgnet.com/1007-9327/full/v11/i1/118.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i1.118
Small molecules, such as steroids, heme byproducts, free fatty acids, environmental contaminants, xenobiotics, drugs and dietary byproducts, can be efficiently eliminated via the addition of gulcuronic acids. This process is called glucuronidation and catalyzed by uridine diphosphate glucuronosyltransferases (UGTs).
UGTs are important enzymes in phase II metabolism, which are located on the luminal side of the endoplasmic reticulum and in the nuclear envelope of cells of the liver and other organs. There are many isoforms of UGTs. To date, three UGT families have been identified: UGT1, UGT2 and UGT8[1], and at least thirteen different UGT1 genes have been characterized in human beings[2].
In human beings, UGT1A3 and UGT1A4 are the only two enzymes which can catalyze N+-glucuronidation of tertiary amines[3,4], including many important drugs in clinic such as imipramine[5], olanzapine[6], ketotifen[7], etc. In addition, UGT1A3 can also catalyze O-glucuronidation of opioids, coumarins, flavonoids, anthraquinones and small phenolic compounds[8]. Drugs containing a carboxylic acid moiety, such as nonsteroidal anti-inflammatory agents and fibrates, are substrates for human UGT1A3[9,10].
The glucuronidation is so important that its disorders can cause drug-induced adverse reactions, variations of cancer susceptibility and many serious diseases[11-13], such as Crigler-Najjar syndrome type I (CN-I), neonatal hyperbilirubinemia. Opioids and nicotine addiction may also be influenced by glucuronidation[14]. Recently, it has been proved that glucuronidation represents a mechanism of intrinsic drug resistance in colon cancer[15]. In fact, most disorders of gulcuronidation are due to dysfunction of UGTs[16,17]. Studies on UGTs are important not only for the metabolism of small molecules, such as drugs, but also for disease pathogenesis.
In this study, the human gene encoding UGT1A3 was obtained from human livers and finally expressed in CHL cells by reconstructing them into pcDNA3.1 (+). The activity of the recombinant enzyme was also assayed with its flavonoid substrate, quercetin.
Trypsin, minimum essential medium, fetal calf serum, TRIzol reagent and G418 were purchased from Gibco BRL (Grand Island, NY, USA). M-MuLV reverse transcriptase, restriction enzymes, DNA molecular marker and T4 ligase were obtained from MBI Fermentas (Amherst, NY, USA). Agarose LE was supplied by Roche Diagnostics (Mannheim, Germany). Uridine 5’-diphospho-glucuronic acid (UDPGA), Brij58, d-saccharic acid 1,4-lactone, β-glucuronidase and diethylprocarbonate were provided by Sigma Chemical Co. (St. Louis, MO, USA). pGEM-T vector was from Promega (Madison, WI, USA). Quercetin and morin were purchased from National Institute for the Control of Pharmaceutical and Biological Products, pcDNA3.1 (+) was from Invitrogen (Calsbad, CA, USA). Other biochemical and molecular biology reagents were obtained from Sangon (Shanghai, China). All other reagents and organic solvents of analytical or HPLC grades were commercially available.
CHL cells (maintained by Department of Pathology and Pathophysiology, College of Medicine, Zhejiang University) were cultured in minimum essential medium containing 100 mL/L fetal calf serum, 0.3 g/L of l-glutamine, 100000 U/L penicillin and 100 mg/L streptomycin. Cells were grown at 37 °C in a humidified atmosphere containing 50 mL/L CO2.
This study was approved by the Ethics Committee of Zhejiang University. Two liver samples were obtained from two Chinese patients at the First Affiliated Hospital of Zhejiang University. One gastric tissue sample was from Zhejiang Provincial Tumor Hospital. In all instances, samples were performed at the distal resection margin of the specimen and exhibited no signs of macroscopic deterioration. All tissues were immediately frozen in liquid nitrogen and stored at -80 °C until use.
The sequence corresponding to UGT1A3 was produced by RT-PCR using gene-specific primers. Total RNA was extracted from human tissue samples with TRIzol reagent following the manufacturer’s instructions. Reverse transcription and amplification were performed by M-MuLV reverse transcriptase and Pfu DNA polymerase, respectively. The sense primer for PCR was 5’-aagcttgaagaaagcaaacgtagcaggc-3’, corresponding to the nucleotides -65 to -44 of UGT1A3. A T in -53 was mutated to C to avoid an interferential start codon, and a Hind III site was introduced to the beginning. The antisense primer was 5’-ctcgagtaccttatttcccacccacttc-3’, with a Xho I site at 5’ side. PCR was performed at 94 °C for 2 min, then 32 cycles at 94 °C for 20 s, at 56.2 °C for 30 s, at 72 °C for 2 min, and a final extension at 72 °C for 10 min. Amplified gene was sequenced after ligation with a pGEM-T vector.
Hind III and Xho I sites were used to introduce UGT1A3 gene into the mammalian expression vector pcDNA3.1 (+). The recombinant plasmid was transformed into E. coli strain DH5α. After screened by ampicillin, the recombinants were identified by restriction enzyme digestion.
The correct recombinant was transfected into CHL cells at 70-80% confluency using a calcium phosphate method. Concentration of G418 was kept at 400 mg/L in medium in the first selection passage to eliminate the cells that failed to be transfected until untransfected cells in control group were completely killed. G418 was added at 200 mg/L serving for maintaining resistant cells in the later passages. The selection concentrations were determined by preliminary experiments according to the susceptibility of CHL cells to G418. After selection, surviving cells were diluted and inoculated into 96-well plates to obtain resistant colonies. Several resistant colonies were harvested and cultured in medium containing G418 respectively to produce UGT1A3 protein.
Preparation of S9 of CHL-UGT1A3 was in the same way reported previously[18] except for three freeze-thaw cycles before sonication. In brief, cells were washed twice with PBS and scraped into 11.5 g/L KCl. After three freeze-thaw turns, cells were sonicated five times, 3 s each time, with bursts for 5 s on ice. The supernatant was obtained by centrifuging for 20 min at 9000 g. The concentration of protein was determined by the method of Lowry, and the rest proteins were stored at -80 °C until use.
Transcription of UGT1A3 in CHL cells was confirmed by RT-PCR. Primers of β-actin were also added into PCR mixture to assure the feasibility of PCR reaction system. Control reactions using total RNA of untransfected CHL cells as templates were performed in parallel.
A typical incubation mixture (100 μL of total volume) contained 100 mmol/L Tris-HCl buffer, pH 7.5, 10 mmol/L MgCl2, 5 mmol/L UDPGA, 5 mg/L Brij58, 1 g/L S9, 5 mmol/L d-saccharic acid 1,4-lactone and 0.4 mmol/L quercetin. The mixture without UDPGA was preincubated at 37 °C for 2-3 min. Reaction was initiated by the addition of UDPGA and incubated at 37 °C for an appropriate time, and then terminated with 290 μL of methanol. After addition of an internal standard (0.4 mmol/L Morin in mixture), the mixture was centrifuged to remove precipitated protein, and 25 μL of supernatant was subjected to HPLC analysis. Blank incubations without UDPGA or S9 or quercetin were performed, and S9 of untransfected CHL cells was also treated simultaneously. The glucuronide of quercetin was confirmed by UV scanning with a DAD detector and hydrolysis with β-glucuronidase.
Analysis was performed on a Shimadzu LC-10A (Kyoto, Japan) system, equipped with two LC-10AD pumps, a DAD UV detector and a DiamonsilTM C18 column (5 μm particle size, 250 mm×4.6 mm). Data acquisition and integration were performed using a HS2000 chromatography workstation. The mobile phase consisted of 540 mL/L methanol and 460 mL/L of 0.02 mol/L phosphoric acid, and flow velocity was 1 mL/min. Quercetin and its metabolites were detected at 368 nm.
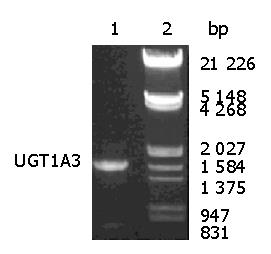
The sequence obtained from RT-PCR was identical with human UGT1A3-3 allele (GenBank accession nos. AY435138 and AF465194) containing three single nucleotide polymorphisms(SNPs) compared with wild type. Figure 1 shows the PCR products electrophoresed on 8 g/L agarose gel. The image was scanned and analyzed by gel image system (Bio-Rad Laboratories, Segrate, Italy).
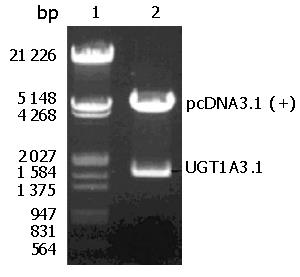
Plasmid pcDNA3.1 (+)-UGT1A3 was extracted from E. coli DH5α. After the recombinant was digested with Xho I and Hind III in combination, a fragment of 1600 bp and a fragment of pcDNA3.1 (+) vector were observed simultaneously on 8 g/L agarose gel (Figure 2). The image was scanned and analyzed by gel image system (Bio-Rad Laboratories, Segrate, Italy).
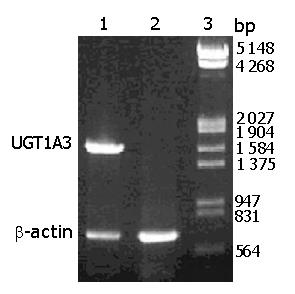
The transcription of UGT1A3 in CHL-UGT1A3 is shown in Figure 3. RT-PCR using total RNA of CHL-UGT1A3 cells as templates showed a fragment about 1 600 bp, and a β-actin fragment (about 660 bp) was synthesized in the reaction. Control reaction corresponding to untransfected CHL cells could synthesize only one β-actin fragment.
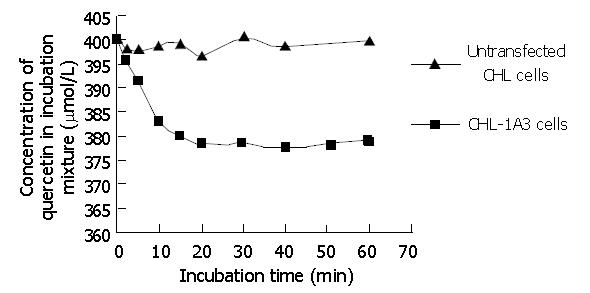
The concentration of S9 obtained from recombinants was 4.3±1.2 mg/mL (n = 4). For analysis, a standard curve was prepared by plotting S (S = peak area of quercetin / peak area of morin) versus the concentration of quercetin (μmol/L). The linear regression of standard curve was determined to be Y = 4.7×10-3X +2.9×10-3(r2 = 0.99). Preliminary experiments also indicated that the glucuronidation reaction was linear for up to 10 min incubation (Figure 4), and the methods used here had satisfactory accuracy and precision.
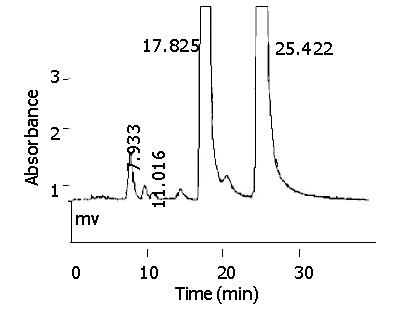
After incubation, quercetin-glucuronide, morin and quercetin were eluted at retention times of about 7.9 min, 17.8 min and 25.4 min, respectively (Figure 5). A uv scanning study by DAD indicated quercetin and its metabolites shared the similar uv absorption spectra, and the metabolite peak disappeared in chromatography after hydrolysis with β-glucuronidase. Blank incubations without UDPGA or S9 or quercetin and the control reaction with S9 of untransfected CHL cells showed no signal of metabolites.
In this work, one active UGT1A3 protein was obtained in CHL cells. But sequence analysis indicated its gene was an allele (UGT1A3-3) of wild UGT1A3. It has three point mutations. Two of them are sense mutations, and the rest one is a silent mutation. This allele was found in all three individual tissue samples, including two liver tissues and one gastric tissue at different times. We consider there are two possibilities: one is that the mutations might be created by PCR reactions, the other is that the frequency of UGT1A3-3 allele might be high in Chinese population, which is a challenging question to the prevalence of UGT1A3 alleles in Chinese population. A recent study reported that the two sense mutations in this allele increased the activity of UGT1A3 enzyme[19], but its clinical significance remains to be explored.
In the past, most studies on drug metabolic enzymes were based on animal experiments, human liver microsomes or enzymes purified from human tissues[20-22]. They are all efficient but have obvious flaws. Human liver microsomes are good models for drug metabolic enzyme studies, but their rare resources and the complexity of mixed enzymes in microsomes limit their applications. Animal experiments are held back by the problem of species differences, and the fact that UGTs are proteins on membranes determines the difficulties in process. In recent years, the use of enzyme preparations expressed in heterologous expression system has become more popular for the studies on drug metabolic enzymes because of their purity and constant supply. Some heterologous expression systems, such as bacteria[23,24], mammalian cells and insect cells are used to express UGTs[25,26]. Mammalian cells have several advantages compared to other expression systems in terms of perfect post-translational modifications, easy to use and low cost to maintain. Although there are studies on the expression of UGTs in some mammalian cells[27-29], to our knowledge, there has been no report on the expression of active UGT1A3 in CHL cells.
Flavonoids are polyphenolic compounds that occur ubiquitously in plants and human diets. Quercetin belongs to the class of water-soluble plant pigments. There is evidence that quercetin possesses potent antioxidant properties[30]. It protects low density lipoprotein cholesterol from becoming damaged and is considered as a crucial intermediate in the formation of atherosclerotic plaques[31]. Quercetin has been reported to be an effective substrate of UGT1A3[8]. In this experiment, only one glucuronide of quercetin was detected after catalysis of recombinant UGT1A3, although it was reported that four quercetin monoglucuronides corresponding to four hydroxyl groups (3-, 7-, 4’-, 3’-, respectively), were produced by incubation with human liver cell-free extracts[32] or human UGT-1A9 microsomes. It could be explained as the limits of recombinant enzyme activity or detection sensitivity, but there is still another possibility that glucuronidation of quercetin catalyzed by UGT1A3 is region-selective. Only one hydroxyl group could form glucuronides in such conditions. Further work should be done to make it clear. For this purpose, more effective expression systems and more sensitive activity detection technologies need to be developed.
In conclusion, active human UGT1A3 protein is expressed in CHL cells, and its activity can be assayed with quercetin. It may provide convenience to related studies on drug metabolisms and mechanisms of diseases.
Co-first-authors: Ya-Kun Chen
Edited by Kumar M and Wang XL
| 1. | Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1179] [Cited by in RCA: 1107] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 2. | Gong QH, Cho JW, Huang T, Potter C, Gholami N, Basu NK, Kubota S, Carvalho S, Pennington MW, Owens IS. Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics. 2001;11:357-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 219] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 3. | Kurkela M, García-Horsman JA, Luukkanen L, Mörsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278:3536-3544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 126] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 4. | Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31:1361-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Nakajima M, Tanaka E, Kobayashi T, Ohashi N, Kume T, Yokoi T. Imipramine N-glucuronidation in human liver microsomes: biphasic kinetics and characterization of UDP-glucuronosyltransferase isoforms. Drug Metab Dispos. 2002;30:636-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Linnet K. Glucuronidation of olanzapine by cDNA-expressed human UDP-glucuronosyltransferases and human liver microsomes. Hum Psychopharmacol. 2002;17:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Breyer-Pfaff U, Mey U, Green MD, Tephly TR. Comparative N-glucuronidation kinetics of ketotifen and amitriptyline by expressed human UDP-glucuronosyltransferases and liver microsomes. Drug Metab Dispos. 2000;28:869-872. [PubMed] |
| 8. | Green MD, King CD, Mojarrabi B, Mackenzie PI, Tephly TR. Glucuronidation of amines and other xenobiotics catalyzed by expressed human UDP-glucuronosyltransferase 1A3. Drug Metab Dispos. 1998;26:507-512. [PubMed] |
| 9. | Sabolovic N, Magdalou J, Netter P, Abid A. Nonsteroidal anti-inflammatory drugs and phenols glucuronidation in Caco-2 cells: identification of the UDP-glucuronosyltransferases UGT1A6, 1A3 and 2B7. Life Sci. 2000;67:185-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Ghosal A, Hapangama N, Yuan Y, Achanfuo-Yeboah J, Iannucci R, Chowdhury S, Alton K, Patrick JE, Zbaida S. Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia). Drug Metab Dispos. 2004;32:314-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 96] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | de Leon J. Glucuronidation enzymes, genes and psychiatry. Int J Neuropsychopharmacol. 2003;6:57-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 12. | Francoual J, Trioche P, Mokrani C, Seboui H, Khrouf N, Chalas J, Clement M, Capel L, Tachdjian G, Labrune P. Prenatal diagnosis of Crigler-Najjar syndrome type I by single-strand conformation polymorphism (SSCP). Prenat Diagn. 2002;22:914-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Wells PG, Mackenzie PI, Chowdhury JR, Guillemette C, Gregory PA, Ishii Y, Hansen AJ, Kessler FK, Kim PM, Chowdhury NR. Glucuronidation and the UDP-glucuronosyltransferases in health and disease. Drug Metab Dispos. 2004;32:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 176] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 14. | Tricker AR. Nicotine metabolism, human drug metabolism polymorphisms, and smoking behaviour. Toxicology. 2003;183:151-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Cummings J, Ethell BT, Jardine L, Boyd G, Macpherson JS, Burchell B, Smyth JF, Jodrell DI. Glucuronidation as a mechanism of intrinsic drug resistance in human colon cancer: reversal of resistance by food additives. Cancer Res. 2003;63:8443-8450. [PubMed] |
| 16. | Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3:136-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 291] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 17. | Duguay Y, McGrath M, Lépine J, Gagné JF, Hankinson SE, Colditz GA, Hunter DJ, Plante M, Têtu B, Bélanger A. The functional UGT1A1 promoter polymorphism decreases endometrial cancer risk. Cancer Res. 2004;64:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 18. | Li X, Yu YN, Zhu GJ, Qian YL. Cloning of UGT1A9 cDNA from liver tissues and its expression in CHL cells. World J Gastroenterol. 2001;7:841-845. [PubMed] |
| 19. | Iwai M, Maruo Y, Ito M, Yamamoto K, Sato H, Takeuchi Y. Six novel UDP-glucuronosyltransferase (UGT1A3) polymorphisms with varying activity. J Hum Genet. 2004;49:123-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Kim KA, Lee JS, Park HJ, Kim JW, Kim CJ, Shim IS, Kim NJ, Han SM, Lim S. Inhibition of cytochrome P450 activities by oleanolic acid and ursolic acid in human liver microsomes. Life Sci. 2004;74:2769-2779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Hu YZ, Yao TW. In vitro metabolism and inductive or inhibitive effect of DL111 on rat cytochrome P4501A enzyme. Chem Biol Interact. 2004;147:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Hutchinson MR, Menelaou A, Foster DJ, Coller JK, Somogyi AA. CYP2D6 and CYP3A4 involvement in the primary oxidative metabolism of hydrocodone by human liver microsomes. Br J Clin Pharmacol. 2004;57:287-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 91] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 23. | Fujita K, Mogami A, Hayashi A, Kamataki T. Establishment of Salmonella strain expressing catalytically active human UDP-glucuronosyltransferase 1A1 (UGT1A1). Life Sci. 2000;66:1955-1967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 24. | Coffman BL, Kearney WR, Green MD, Lowery RG, Tephly TR. Analysis of opioid binding to UDP-glucuronosyltransferase 2B7 fusion proteins using nuclear magnetic resonance spectroscopy. Mol Pharmacol. 2001;59:1464-1469. [PubMed] |
| 25. | Goto A, Adachi Y, Inaba A, Nakajima H, Kobayashi H, Sakai K. Identification of human p450 isoforms involved in the metabolism of the antiallergic drug, oxatomide, and its inhibitory effect on enzyme activity. Biol Pharm Bull. 2004;27:684-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Kaku T, Ogura K, Nishiyama T, Ohnuma T, Muro K, Hiratsuka A. Quaternary ammonium-linked glucuronidation of tamoxifen by human liver microsomes and UDP-glucuronosyltransferase 1A4. Biochem Pharmacol. 2004;67:2093-2102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Turgeon D, Chouinard S, Belanger P, Picard S, Labbe JF, Borgeat P, Belanger A. Glucuronidation of arachidonic and linoleic acid metabolites by human UDP-glucuronosyltransferases. J Lipid Res. 2003;44:1182-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 28. | Cuff RL, Wade LT, Rychlik B, Jedlitschky GA, Burchell B. Characterisation of glucuronidation and transport in V79 cells co-expressing UGT1A1 and MRP1. Toxicol Lett. 2001;120:43-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Jinno H, Saeki M, Saito Y, Tanaka-Kagawa T, Hanioka N, Sai K, Kaniwa N, Ando M, Shirao K, Minami H. Functional characterization of human UDP-glucuronosyltransferase 1A9 variant, D256N, found in Japanese cancer patients. J Pharmacol Exp Ther. 2003;306:688-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Milane HA, Ubeaud G, Vandamme TF, Jung L. Isolation of quercetin's salts and studies of their physicochemical properties and antioxidant relationships. Bioorg Med Chem. 2004;12:3627-3635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 31. | Hollman PC, Katan MB. Absorption, metabolism and health effects of dietary flavonoids in man. Biomed Pharmacother. 1997;51:305-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 388] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 32. | Day AJ, Bao Y, Morgan MR, Williamson G. Conjugation position of quercetin glucuronides and effect on biological activity. Free Radic Biol Med. 2000;29:1234-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 258] [Article Influence: 10.3] [Reference Citation Analysis (0)] |