MATERIALS AND METHODS
H pylori culture and preparation of VacA+ BCS and VacA- BCS
After detection of their morphology and biochemical response and identification of the genotype of 10 clinically-derived H pylori strains; 00-1466 and 00-1783 with the genotype (s1a/m1/cagA+) and (s1b/m2/cagA+) respectively were used in the following experiment as VacA+ and VacA- challenge factors separately. Then the two strains were cultured in brain heart infusion (BHI) broth +10% fetal bovine serum (FBS) in mixed air containing 5% O2, 10 mL/L CO2, 80% N2 at 37 °C with agitation (200 r/min) for 48 h. Incubation was terminated when the turbidity value reached McFarland 6-7 (about 3-9×108 CFU/mL). Gram staining was done in order to assure no contamination of other bacteria. The cultures were centrifuged at 15000 g for 30 min at 4 °C and the supernatant was filtrated with 0.2 mm syringe filter. The sterile supernatant was stored at -20 °C.
VacA+ BCS infection of SGC7901 cells
Cell concentration was adjusted to 5×105/mL after digested with 0.05% trypsine. Then these cells were cultured in six 25 cm2 cell flasks and incubated at 37 °C for 24 h. The culture medium was discarded and cells were rinsed with DMEM containing no fetal calf serum (FCS). Six flasks were equally divided into two groups. VacA+ BCS derived from 00-1466 and VacA- BCS derived from 00-1783 were added to the different flasks as positive and negative controls respectively, and then cells were incubated at 37 °C for 6 h.
Total RNA isolation and poly (A)+ mRNA preparation
After incubation for 6 h, Vac A+ BCS was discarded and cells were rinsed with DMEM containing no FCS. Total RNA was isolated from cells using TRIZOL regents (GIBCO BRL) to prepare cDNA probes. RNA quality was assessed using 1.2% agarose gel electrophoresis in the presence of ethidium bromide. Samples that failed to reveal intact 18S and 28S ribosomal bands were excluded for further study. Poly (A) + mRNA was isolated from the total RNA using a poly (dT) resin (Qiagen, Hilden, Germany). In order to adjust the differences in probe intensity distribution across different chips, 8 house-keeping genes were used as inner control.
RT-PCR and preparation of 33P-labelled cDNA
Approximately 1-2 ug of mRNA was labeled in a reverse transcription reaction in the presence of 200 mCi [a-33P]deoxyadenosine 59-triphosphate (DuPont NEN, Boston, MA) using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s instructions (Promega Corp., Madison, WI). For RT-PCR, 1% of the reverse transcription reaction was amplified using Taq DNA polymerase for 30 s at 94 °C, for 1 min at 55 °C, and for 1.5 min at 72 °C for 35 cycles.
Hybridization and image procession
Prehybridization was carried out in 20 mL of prehybridization solution (6×SSC, 0.5% SDS, 5XDenhardt’s, and 100 μg/mL denatured salmon sperm DNA) at 68 °C for 3 h. Overnight hybridization with 33P-labeled cDNA in 6 mL of hybridization solution (6× SSC, 0.5% SDS, and 100 μg/mL salmon sperm DNA) was followed by stringent washing (0.1×SSC, 0.5% SDS, 65 °C for 1 h). Membranes were exposed to phosphor screen overnight and scanned using a FLA-3000A plate/fluorescent image analyzer (Fuji Photo Film, Tokyo, Japan). Radioactive intensity of each spot was linearly digitalized to 65536 gray-grade in a pixel size of 50 mm in an image reader and recorded using the Array Gauge software (Fuji Photo Film, Tokyo, Japan). After subtraction of background (3±3) chosen from an area where no cDNA was spotted, genes with intensities>10 were considered as positive signals to ensure that they were distinguished from background with statistical significance >99.9%. Normalization among arrays was based on the sum of background-subtracted signals from all genes on the membrane.
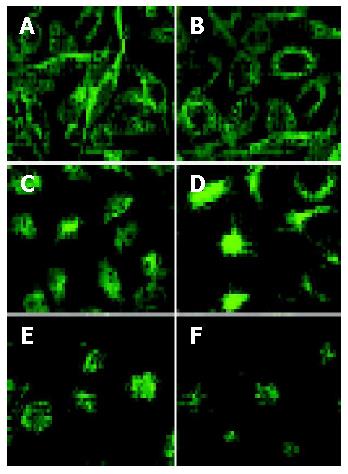
Effect of VacA BCS on cytoskeleton of SGC7901 and HeLa cells
SGC7901 and HeLa cells were cultured on a 6-well plate with a cover slide in each well. After incubation at 37 °C with 5 mL/L CO2 for 36 h, the culture medium was substituted with 1 mL VacA-BCS or VacA+BCS and 1ml culture medium. Cells were harvested at 12, 18, 24, 36 and 48 h. After washing 3 times with PBS, cells were fixed in 2 mL 4% citromint at room temperature for 20 min, and washed with PBS 3 times, and 0.3% Triton X-100 was added. Following incubation at room temperature for 15 min and washing with PBS, 1 mL 5% rabbit serum was overlaid, cells were incubated at 4 °C overnight, and washed again and a monoclonal anti-tubulin antibody (Oncogene) was added. Following incubation at 37 °C for 1 h and washing, anti-mouse IgG (DAKO Danmark) was added, the slides were washed and cytoskeleton change was observed through a high-resolution laser scanning confocal microscopy (LSCM).
DISCUSSION
The cDNA micoarray technology can screen thousands of genes responsive to some virulence determinants simultaneously and identify host gene expression patterns, during which the relationship between a virulence determinant and regulated genes of host cells may be established. It was used in the present study to examine the transcriptional response of gastric epithelial SGC7901 cells to VacA BCS in vitro.
H pylori infection could result in various diseases such as chronic gastritis, peptic ulcer and gastric carcinoma[4]. It is helpful to understand the biological process and the pathogenesis of H pylori-associated diseases and the role of VacA in the pathogenicity of H pylori by finding out gene expression alterations in cells challenged by H pylori. In this study, target cells were always incubated with H pylori or its components to observe the biological changes. After co-incubation, target cells often had growth arrest, vacuolation, apoptosis, cytokine secretion, and cytoskeleton rearrangement[5-8]. Gene chip technique was used to clarify the biological response of cells to H pylori or its toxic components and the mechanism of H pylori-related diseases. The results showed that there were 198 genes with 3 fold different expression levels and 865 genes with 2 fold different expression levels in cells challenged by VacA+ BCS. Genes with a distinct expression difference account for about 5% of total genes[9-11] and those with a definite function are involved in many important cell processes. These genes include oncogene, tumor suppressor gene, ion channel transportation associated gene, gene associated with stress response and inflammation, cell cycle-related gene, cytoskeleton-associated gene and gene related to cell apoptosis.
Expression alteration of cytoskeleton associated genes
VacA is able to delay healing of gastric ulcer in rats, inhibit reepithelialization, and worsen the quality of mucosa scar in vivo[12], inhibit in vitro gastric epithelial cell proliferation and interfere with EGF-induced signal transduction essential for healing of gastric mucosa[13]. VacA could disrupt cytoskeletal architecture essential for the maintenance of cell structure integrity and epithelial barrier function[14]. Studies by Ashorn et al[15] favored the role of VacA in cytoskeletal rearrangements, because cytotoxin-produed H pylori strains could lead to condensation of filamentous actin, while cytotoxin-negative strains did not cause marked disturbances in the intracellular structure of cells. Cell vacuolation is associated with cytoskeletal rearrangements[16,17], VacA-mediated cell vacuolation is strongly correlated to the inhibition of reepithelialization and loss of stress fibers[18].
Cytoskeleton is composed of microfilaments, microtubules, and intermediate filament cross-linked bundles, which are anchored to other cellular components, including cell membrane. Microtubules are important cytoplasmic structures involved in intracellular transport and are essential for cell division and differentiation. Intermediate filaments consist of fibrous protein, which has been shown to have mechanical functions in stiffening cells and organizing intracellular organelles for coordinated activity[14]. Our microarray analysis indicates cytoskeleton-related genes are expressed at low levels, including kinesin-like-5 protein, actin-related protein 2 (ARP2), intermediate filament protein syncoilin and dynactin 4 (Table 1). Kinesin-like-5 protein is a member of the kinesin-like protein superfamily, specifically regulates microtubule formation and consecutive movement of chromosomes during mitosis, and therefore is crucial for cell division. In recent years, actin-related protein 2/3 (Arp2/3) complex has emerged as a central effector of actin assembly that receives multiple signal inputs inducing the formation of branched filaments. The Arp2/3 complex is composed of 7 evolutionarily conserved subunits: 2 actin-related proteins (Arp2 and Arp3) and 5 other subunits, which in yeast are called Arc40, Arc35, Arc18, Arc19, and Arc15. In all organisms examined, the Arp2/3 complex is localized at the site of dynamic actin assembly. In yeast, the Arp2/3 complex is localized at the cortical actin patches, highly motile filamentous actin structures. Mutations in different subunits of the yeast Arp2/3 complex could disrupt actin organization, actin patch motility, and actin-dependent processes such as endocytosis, cell polarity development, and organelle inheritance[19]. Syncoilin is a 64-kDa protein found in skeletal and cardiac muscles and has been proposed as a member of the intermediate filament (IF) protein superfamily based on sequence analysis. Syncoilin is involved in the anchoring of the desmin intermediate filament network at the sarcolemma and neuromuscular junction that is likely to be important in maintaining muscle fiber integrity and may also link dystrophin-associated protein complex to cytoskeleton. Its dysfunction or absence might result in disruption of the intermediate filament network and muscle necrosis[20]. Dynactin is a complex with at least 10 polypeptides from 24 to 150 ku in size. The best characterized subunits of dynactin are p150Glued and p50 (dynamitin). p150Glued contains both microtubule- and dynein-binding domains. p150Glued could bind to microtubules through the NH2-terminal CAP-Gly motif, and phosphorylation near this motif has been shown to regulate microtubule binding. p150Glued could interact directly with the intermediate chain of cytoplasmic dynein (DIC) that may be essential for dynein-mediated organelle transport. Regulation of this interaction by DIC phosphorylation has suggested an important function[21]. Expression of genes encoding cytoskeleton-associated proteins essential in keeping the normal cell architecture is inhibited after cells are challenged by VacA+ BCS. This decrease results in reduction of the production of proteins encoded by these genes and collapse of cytoarchitecture. This alteration of gene expression is an early response of cells interfered with exotic factors, which precedes morphologic change of cell configuration and is the fundamental cause of cell morphologic change. But further study is still needed to clarify the mechanism of VacA+ BCS to induce cytoskeleton changes and the relationship between morphologic and cytoskeleton changes of cells. Further research on the effect of VacA+ BCS on cell morphology especially on cytoskeleton will be helpful to understand how VacA and other secreted bacterial toxic factors induce cell pathological changes.
Expression change of tumorigenesis-associated genes
H pylori is associated with gastric cancer and MALT-lymphoma. In 1994, H pylori was classified by the International Association for Research against Cancer as a type 1 carcinogen[22]. In this study, some tumor-associated genes were highly expressed, such as melanoma antigen (MAGE) gene, jun B oncogene, homologue of mouse MAT-1 oncogene, annexin A10 gene, telomeric repeat binding factor 2 gene, fibroblast growth factor receptor gene, S phase kinase associated protein (p45) gene. MAGE exists in tumor cells but not in normal cells, and is related to differentiation of tumor cells. It is a symbol of malignant cells[23,24]. Expression of tumor suppressor gene that encodes KIAA0456 protein (Dab2) decreases in 80% oophoro carcinomas. Inactivation or down-expression of Dab2 could probably induce or promote the development of oophoro carcinoma[25]. Our results show that tumor suppressor genes such as Dab2 were up-regulated, which is inconsistent with the fact that VacA+ BCS could result in increased expression of most tumor associated genes. But we found the same thing from other correlative studies, in which the expression level of tumor suppressor genes increased correspondingly with increased expression levels of oncogenes. Several ANXs are involved in tumorigenesis. ANXA1 is overexpressed in breast cancer and hepatocellular carcinoma (HCC). ANXA2 is overexpressed in brain glial tumors and pancreatic cancer and ANXA8 in acute promyelocytic leukemia. Overexpression of ANXA1 correlates positively with metastatic potential of breast cancer and increased growth of tumor cells inoculated in nude mice. In contrast, ANXA6 has tumor suppressive activities in squamous cell carcinoma, and reduction of ANXA6 and ANXA7 proteins is associated with malignant phenotype and the metastatic potential of melanoma. Moreover, ANXA4 plays a role in chemoresistance. ANXA10 may be a potential tumor suppressor gene because its down-regulation is associated with malignant phenotype of liver cells, and vascular invasion and progression of HCC[26]. Proliferation and death are two different physiological behaviors of cells. The equilibration between them is helpful to keep cell stabilization. Disruption of this equilibration would induce cell pathologic changes such as apoptosis or proliferation. VacA+ BCS could enhance these two important potentials of cells, and consequently boost the sensitivity of cells to pathogens.
Telomerase, a reverse transcriptase composed of RNA and proteins, can synthesize telomeric repeat sequence and add this sequence to the terminal of chromosome to keep the eternal life of cells. Activation of telomerase is a biological characteristic of malignant tumor[27]. In this study, increased expression of TP-1, the main component of telomerase, was observed. The expression state of tolemerase in cells challenged by VacA+ BCS still needs to be confirmed. The expression alterations of tumor-related genes in cells challenged by VacA+ BCS indicates that VacA+ BCS has the potential to promote canceration of cells.
Expression alterations of stress response genes and genes encoding inflammation-associated factors
Stress or heat shock proteins (HSPs) are ubiquitous and highly conserved proteins whose expression is induced in response to a wide variety of physiological and environmental insults. They allow cells to survive in otherwise lethal conditions. Some of the important house-keeping functions attributable to the molecular chaperones include: import of proteins into cellular compartments; folding of proteins in cytosol, endoplasmic reticulum, and mitochondria; degradation of unstable proteins; dissolution of protein complexes; prevention of protein aggregation; control of regulatory proteins and refolding of misfolded proteins[28]. Accumulation of abnormally folded proteins in nuclei or cytosol that occurs as a consequence of stress, increases temperature, free oxygen radicals, heavy metals, and even antibiotics[29]. Formation of aggregates could disturb normal cellular function and trigger cell death. Such a mechanism is involved in the pathogenesis of lesions that characterize several neurodegenerative disorders. Most HSPs are involved in the proper folding and/or elimination of misfolded proteins, thus contributing to cell survival. The relationship between HSPs and tumors is reported[30]. The yield of HSPs is increased in canceration and tumor cells[31]. In this study, expression of all HSP genes increased, indicating that abnormal states of target cells and the potential of VacA+ BCS can induce cell canceration.
IL-14 secreted by activated T and B cells can activate division of B cells and promote proliferation of some subgroups of B cells[32]. In this study, decreased expression of IL-14 gene was found to be different from other inflammatory factors. Gene expression level of other inflammation-associated factors increased, such as α-interferon, tumor necrosis factor receptor- associated proteins, interlukin-1 receptor antagonist. These results are coincident with the cell response to H pylori and its metabolic products. The interfering factor in this study was VacA+ BCS. Thus we could deduce that VacA might play an important role in this process.
The results of this study clearly indicate that H pylori VacA+ BCSs can induce expression changes at multi-gene level in infected cells, thus adjusting the expressed products according to the interference with various physiological functions and activities of target cells.
In conclusion, VacA+ BCS can cause alterations of cell gene expression profile at multi-level and multi-phase and such alterations are almost involved in all important pathologic processes of H pylori-associated diseases. It is suggested that VacA plays an important role in the pathogenic mechanism of H pylori.