Published online May 1, 2004. doi: 10.3748/wjg.v10.i9.1286
Revised: October 2, 2003
Accepted: October 27, 2003
Published online: May 1, 2004
AIM: Bcl-2/adenovirus E1B 19 ku interacting protein 2-like (BNIPL-2) is a novel protein recently identified in our laboratory. BNIPL-2 is homologous to human BNIP-2, a potentially proapoptotic protein, and can interact with Bcl-2 and Cdc42GAP and promote apoptosis in BEL-7402 cells. Here we report the gene-expression profile regulated by BNIPL-2 in human hepatocarcinoma Hep3B cells and the analysis of its potential roles in cell apoptosis.
METHODS: BNIPL-2 was overexpressed in Hep3B cells using tetracycline inducible or Tet-on system. Screened by Western blot, the cells with low background and high induction fold of BNIPL-2 were obtained. We performed Atlas human cDNA expression array hybridization on these cells and analyzed the data with Quantarray® software to identify BNIPL-2-regulated genes and their expression profile. RT-PCR was used to confirm the altered expression level of part of genes identified by the Atlas array hybridization.
RESULTS: Fifteen of 588 genes spotted on the Atlas membrane showed altered expression levels in BNIPL-2-transfected Hep3B-Tet-on cells, in which 8 genes involved in cell apoptosis or growth inhibition were up-regulated and 7 genes involved in cellular proliferation were down-regulated following overexpression of BNIPL-2.
CONCLUSION: cDNA array is a powerful tool to explore gene expression profiles under inducible conditions. The data obtained using the cDNA expression microarray technology indicates that BNIPL-2 may play its roles in apoptosis through regulating the expression of genes associated with cell apoptosis, growth inhibition and cell proliferation.
-
Citation: Xie L, Qin WX, He XH, Shu HQ, Yao GF, Wan DF, Gu JR. Differential gene expression in human hepatocellular carcinoma Hep3B cells induced by apoptosis-related gene
BNIPL-2 . World J Gastroenterol 2004; 10(9): 1286-1291 - URL: https://www.wjgnet.com/1007-9327/full/v10/i9/1286.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i9.1286
Bcl-2/adenovirus E1B 19 ku interacting protein 2-like (BNIPL) is recently identified and characterized as an apoptosis-associated protein[1-3]. BNIPL shares 72% homology (46% identity) with BNIP-2[4] and has 2 variants BNIPL-1 and BNIPL-2. Full-length amino acid sequence of BNIPL-1 is shorter by 82 amino acids at N-terminus than that of BNIPL-2. Our previous experiment showed that BNIPL-1 could inhibit MIF-mediated proliferation of BEL-7402 cells and BNIPL-2 could inhibit cell growth and promote apoptosis in BEL-7402 cells. However, the molecular mechanisms by which BNIPL-1 suppresses the cell growth and BNIPL-2 commits the cell to die are not known[1,2].
In 1992, Gossen and Bugard developed the tetracycline (or doxycycline)-induced gene expression (Tet-on) system, which is useful for controlling the expression of targeted genes in a quantitative manner and for determining the roles of gene products in cellular functions[5,6]. In the present study, Hep3B-Tet-on cells were transfected with BNIPL-2 using lipofectamine transfection reagent. In order to obtain cells with low background and high induction fold of BNIPL-2, transfected cells were screened by Western blot. Using this system, we established Hep3B cell line with doxycycline-regulated BNIPL-2 expression.
The microarray technique was first reported in 1995 by Schena et al.[7] and allows simultaneous parallel expression analysis of thousands of genes. Information provided by cDNA microarray analysis might be useful for tumor classification, elucidation of key factor in tumors, and identification of genes which might be applied to diagnostic purposes or as therapeutic targets[8,9]. The Atlas human cDNA expression system provides a convenient and quick method for profiling the expression of 588 genes at the same time. In this study, we used cDNA expression microarray technology to analyze the differentially expressed genes regulated by cell apoptosis-related gene BNIPL-2.
cDNA microarray: Atlas human cDNA expression array membranes (#7740-1) were purchased from Clontech Laboratories Inc (Palo Alto, California, United States). The membrane contained 10 ng of each gene-specific cDNA from 588 known genes and 9 housekeeping genes. Several plasmid and bacteriophage DNAs and blank spots were also included as negative and blank controls to confirm hybridization specificity. The 588 known genes spotted on the Atlas memberane consist of cDNAs for cell-cycle control proteins, stress response proteins, apoptosis-associated proteins, DNA transcription factors, cell receptors, extracellular cell signaling and communication proteins, etc. A complete list of the genes with the array positions and GeneBank accession numbers spotted on the array is available at Clontech’s web site (http://www. clontech.com).
Restriction endonucleases BamHI and ClaI were purchased from New England Biolabs Inc (Beverly, Maryland, United States). Lambda DNA/HindIII Markers were from Sino-American Biotechnology Company (Shanghai, China). Tet system approved fetal bovine serum, doxycycline, hygromycin, pTRE2hyg, and Atlas human cDNA expression array trial kit were from BD Biosciences Clontech (Palo Alto, California, United States). Lipofectamine and SuperScriptTM first strand synthesis system for RT-PCR were from InvitrogenTM Life Technologies (Carlsbad, California, United States). High glucose Dulbecco’s modified Eagle’s medium (DMEM) was from Gibco/BRL (Carlsbad, California, United States). Monocolonal antibody actin pan Ab-5, horseradish peroxidase-coupled second antibodies anti-rabbit IgG, and anti-mouse IgG were from Santa Cruz Biotechnology (Santa Cruz, California, United States). SuperSiganl west femto maximum sensitivity substrate and T-PERTM tissue protein extraction reagent was from Pierce Chemical Company (Rockford, Illinois, United States). PROTRAN nitrocellulose transfer membrane was from Schleicher & Schuell (Keene, New Hampshire, United States). Hep3B-Tet-on cell line was constructed by Wang et al[10].
Plasmid construction: For construction of plasmid pTRE2hyg-BNIPL-2, human BNIPL-2 DNA fragment encoding the full-length protein was amplified by PCR using the primers containing BamHI and ClaI restriction sites and HA-tag sequence (Table 1). The PCR product was subcloned into the Tet-on expression vector pTRE2hyg which contains hygromycin-resistance gene. The recombinant plasmid construct, pTRE2hyg-BNIPL-2, was confirmed by BamHI/ClaI digestion and DNA sequencing.
| Gene | GenBank accession number | Orientation | Sequence |
| BNIPL-2 | AY033000 | Forward | 5’ CGGGATCCACCATGTACCCATACGATGTTCCAGATTACGCTCTTGGAACTATACAAGAG 3’ |
| Reverse | 5’ CCATCGATCTATGTCCCTCCTGAGCCATGGAGATCCCGGTCCAGCTGTCTGACAGC 3’ | ||
| β-actin | NM_001101 | Forward | 5’ AAGTACTCCGTGTGGATCGG 3’ |
| Reverse | 5’ TCAAGTTGGGGGACAAAAAG 3’ | ||
| Rad | L24564 | Forward | 5’ GCTGCCCTGCGAAGTGGCTC 3’ |
| Reverse | 5’ TCCCTCCAGGGCAGGCACAC 3’ | ||
| CLK-1 | L29222 | Forward | 5’ GGGTGGTCCCAACCATGTGA 3’ |
| Reverse | 5’ CCGGCAGAACTGTGTTCATCCCA 3’ | ||
| p38 MAP kinase | L35253 | Forward | 5’ GTTGGAACCCCAGGGGCTGA 3’ |
| Reverse | 5’ GGCATGTGCAAGGGCTTGGG 3’ | ||
| p21 | U09579 | Forward | 5’ CTGTGGGGGTGAGGGTCCCA 3’ |
| Reverse | 5’ TGGGGAGGGATGGGGTGGAT 3’ |
Hep3B-Tet-on cells were cultured in DMEM containing 150 mL/L tetracycline-free fetal bovine serum at 37 °C in a 50 mL/L CO2 incubator. They were grown in 6-well plates to 70% confluence and transfected with pTRE2hyg-BNIPL-2 plasmid DNA using lipofectamine reagents under conditions recommended by the manufacturer. After 48 h, the medium was replaced with fresh DMEM containing 100 mg/L hygromycin to select the stable cell lines. Two weeks later, hygromycin-resistant clones were obtained. Thirty-seven clones were selected and cultured continuously. After further selection in the medium containing 25 mg/L hygromycin for more than 3 wk, they were analyzed by Western blot using anti-BNIPL-2 polyclonal antibody (the rabbit anti-BNIPL-2 polyclonal antibody was prepared by National Laboratory for Oncogenes and Related Genes, Shanghai Cancer Institute, Shanghai, China). BNIPL-2 expression in the cell line was induced by incubating cells with doxycycline (Dox), which is a tetracycline analogue.
Cells were washed twice with ice-cold PBS and lysed with T-PERTM tissue protein extraction reagent. Equal amounts of protein (10 μg) were electrophoresed on 100 g/L SDS–PAGE and blotted to PROTRAN nitrocellulose transfer membrane by a semi-dry transfer unit (Amersham Pharmacia Biotech AB, Piscataway, New Jersey, United States). The transferred membranes were blocked with 50 g/L non-fat milk in PBST buffer (20 mmol/L Tris-HCl, pH 7.6; 130 mmol/L NaCl, 1 g/LTween 20) for 1 h at room temperature and subsequently incubated with the primary antibody anti-BNIPL-2 at 1:500 dilutions in PBST containing 40 g/L non-fat milk for 2 h. After several washes with PBST, the membranes were incubated with the corresponding horseradish peroxidase-coupled secondary antibody anti-rabbit IgG in the same buffer. The immunocomplexes were visualized using SuperSignal west femto maximum sensitivity substrate. For standardization and quantification, β-actin was used as an internal control.
Hep3B-Tet-on-BNIPL-2/13 cells were cultured in DMEM containing 150 mL/L tetracycline-free fetal bovine serum at 37 °C in a 50 mL/L CO2 incubator. They were grown in 4 10-cm dishes to 80% confluence and 2 dishes were treated with Dox (2 mg/mL) for 24 h. Then total RNA samples were extracted from 1 × 107 cells using TRIzol reagent. They were reverse-transcribed into cDNA and labeled with [α-32P] dATP. Before labeling, total RNA of each sample was treated with DNase I (TaKaRa, Dalian, Shenyang, China) at 37 °C for 30 min to remove contaminated DNA. For each labeling, the treated 5 μg of total RNA was dissolved in 2 μL of diethyl pyrocarbonate (DEPC)-treated H2O, incubated with 1 μL CDS Primer Mix at 70 °C for 2 min, and continued to incubate at 50 °C for 2 min. Then, 2 μL 5× reaction buffer, 0.5 μL 0.1 mol/L DTT, 1 μL dNTP mixture (containing dCTP, dGTP, and dTTP), 3.5 μL [α-32P] dATP (10 μCi/μL), and 1 μL MMLV reverse transcriptase were added to each sample and incubated at 50 °C for 25 min. The reaction was stopped by adding 1 μL of 10× Termination Mix. The labeled first strand cDNA probes were purified by NucleoSpin extraction spin column to remove the unincorporated nucleotides. After purification, labeled cDNAs were denatured before use.
Five milliliters ExpressHyb hybridization solution and 0.5 mg heat-denatured sheared salmon testes DNA were added to the tube containing Atlas human cDNA expression array membrane, which was pre-hybridized at 68 °C for 3 h. The different probes were then added to different tubes and hybridization was performed at 68 °C for 18 h in a rolling bottle. The membrane was washed twice at 68 °C in 2× SSC, 10 g/L SDS for 20 min, once at 68 °C in 0.1× SSC, 5 g/L SDS for 20 min, and once at room temperature in 2× SSC for 5 min, followed by exposure to x-ray films at -70 °C for 2-5 d.
The images were scanned with CycloneTM storage phosphor system (Packard Bioscience Company, Meriden, Connecticut, United States) and saved as TIFF format files. The TIFF images were imported into the Quantarray® image system (Packard Bioscience Company) and analyzed. Housekeeping genes ubiquitin and β-actin were selected for normalization. The normalized intensity of each spot representing a unique gene expression level was acquired. Genes were considered to be up-regulated when the intensity ratio was > 2 and down-regulated when the intensity ratio was < 0.5.
To confirm the cDNA array results, semi-quantitative RT-PCR was performed for 4 genes (Rad, CLK-1, p38 MAP Kinase and p21) that displayed expression alterations. Five micrograms of total RNA was reverse-transcribed into 20 μL of the first strand cDNA with SuperScriptTM first strand synthesis system, then 2 μL of the product was used as the template to amplify each specific gene fragment in a 25 μL reaction mixture with corresponding primers (Table 1) under the following conditions: denaturation at 94 °C (3 min); 26 cycles at 94 °C (30 s), at 60 °C (30 s), and at 72 °C (30 s); then extension at 72 °C (3 min). In each PCR reaction, primers for the human β–actin gene were used as an internal control. PCR primer pairs were designed using Primer3 Internet software program. Their specificity was confirmed by a BLAST Internet software assisted search for a nonredundant nucleotide sequence database (National Library of Medicine, Bathesda, Maryland, United States). Ten microliters PCR reaction product was analyzed by electrophoresis on a 20 g/L agarose gel and visualized by ethidium bromide staining.
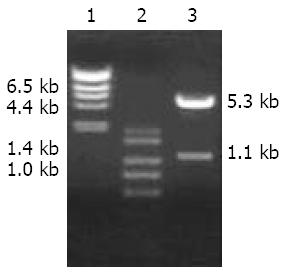
Tet-on expression plasmid pTRE2hyg-BNIPL-2 was successfully constructed. Restriction endonucleases BamHI and Cla I digestion showed that BNIPL-2 gene was correctly inserted into the multicloning site. The BNIPL-2 fragment and pTRE2hyg vector were about 1.1 kb and 5.3 kb, respectively (Figure 1).
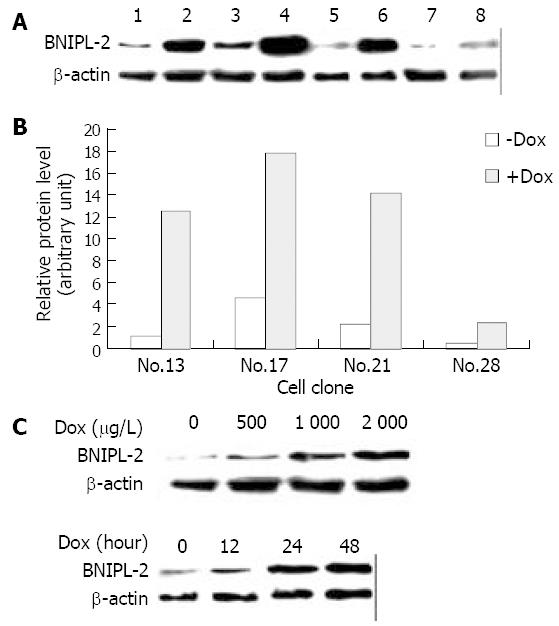
After screening in the medium containing hygromycin for more than 8 wk, a total of 37 independent Hyg-resistant cell lines were obtained from the Hep3B-Tet-on cells, transfected with pTRE2hyg-BNIPL-2. Of the 37 clones, clone Hep3B-Tet-on-BNIPL-2/13 showed a low background and high induction fold expression of BNIPL-2 by Dox and was selected as the cell line for further investigations. The amount of BNIPL-2 induced in the Hep3B-Tet-on-BNIPL-2/13 cells treated with Dox (2000 μg/L) was 12-fold higher than that in the cells without Dox treatment (Figure 2A).
Hep3B-Tet-on-BNIPL-2/13 cells were amplified and passaged (1.0 × 105 cells) into 35-mm dishes, and treated with varying amounts of Dox for different times. Equal amounts of total cell protein were detected by Western blot with BNIPL-2 antibody. The level of BNIPL-2 expression increased significantly after exposing the cells to Dox for 24 h over a range of Dox concentrations from 500 μg/L to 2000 μg/L (Figure 2B). However, the level of BNIPL-2 expression did not increase when Dox concentration was over 2000 μg/L (data not shown). On the other hand, after the cells were treated with Dox (2000 μg/L) for 0, 12, 24, or 48 h, the level of the induction increased with the treatment time and apparent increased induction was seen at 24 h (Figure 2C). Therefore, there was a direct correlation between the concentrations of Dox added to the cells or the time of treatment with Dox and the level of BNIPL-2 induction. The above results obtained by Western blot screening with anti-BNIPL-2 polyclonal antibody were confirmed by anti-HA McAb (data not shown).
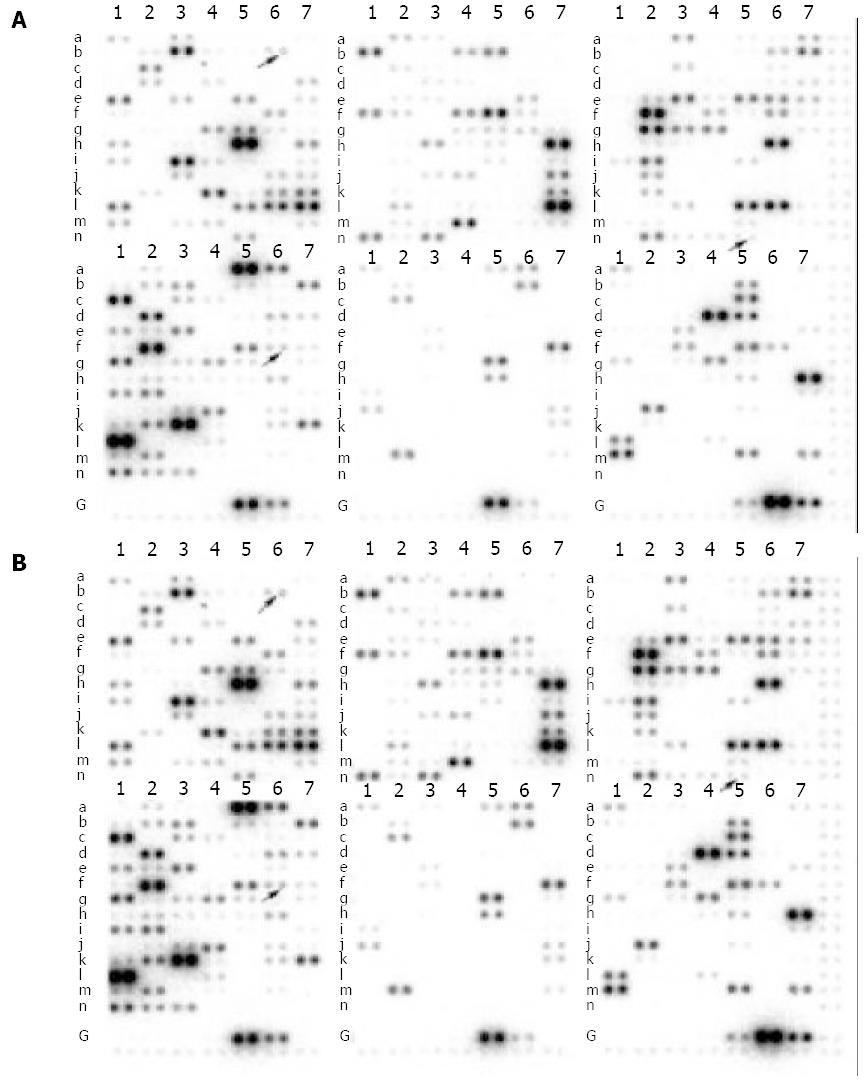
Using a cDNA expression microarray technique we established the expression profile of 588 genes regulated by BNIPL-2 (Figure 3) in Hep3B-Tet-on-BNIPL-2/13 cells. No signals were visible in the blank spots and negative control spots, indicating that Atlas human cDNA array hybridization was highly specific. The density of housekeeping genes was very similar between samples, indicating that the results were reliable. We used the housekeeping genes Ubiquitin and β-actin to normalize the intensities. The comparison results analyzed by Quantarray® image system showed that there were 15 genes altered in terms of their expression levels, of which 8 were up-regulated (Table 2) and 7 were down-regulated (Table 3) following overexpression of BNIPL-2 in Hep3B cells.
| Location | Gene description | Induction-fold |
| C5n | Rad (Ras associated with diabetes) | 5.886 |
| F3e | Insulin-like growth factor binding protein 1 (IGFBP-1) | 2.594 |
| D6f | p21; Cdk-interacting proteins (Cip1); mda-6/WAF1/SDI1 | 2.574 |
| E2m | Interleukin-2 receptor | 2.500 |
| D3n | Transcription factor (E2A) | 2.115 |
| F7m | Estrogen sulfotransferase (EST) | 2.112 |
| F1e | Transforming growth factor-beta (TGF-beta) | 2.018 |
| E7k | lfa-1 alpha subunit | 2.012 |
| Location | Gene description | Induction-fold |
| B4f | Transducin beta-1 subunit 3’end | 0.429 |
| B1b | MAL protein | 0.431 |
| B5f | p38 MAP kinases/CSBP/Mxi2 | 0.442 |
| A7e | Tob | 0.447 |
| A7h | CLK-1 | 0.457 |
| B4b | GTP binding protein (RAB5) | 0.480 |
| A6b | p120 antigen | 0.496 |
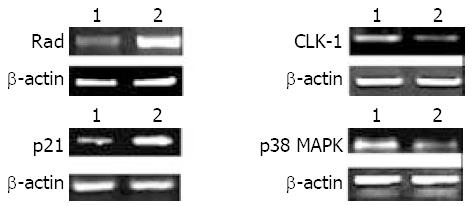
Four genes were measured for their transcript levels by RT-PCR to verify the accuracy and universality of the hybridization data. The RT-PCR results were consistent with hybridization data in each of the genes measured (Figure 4), i.e. Rad and p21 were up-regulated, p38 MAPK and CLK-1 were down-regulated in BNIPL-2 overexpressed cells.
The Tet-on system, which utilizes an E. coli gene regulatory system[11], has been found to have several advantages[12-14]. Namely gene expression is easily regulated by administration of Dox, Dox is minimally toxic, and Dox acts specifically on the target gene, and does not activate other cellular genes. In the present study, we established a cell line Hep3B-Tet-on-BNIPL-2/13 in which BNIPL-2 gene could be expressed conditionally by Dox treatment. Dox activated the Tet-response element and significantly enhanced the expression of BNIPL-2 gene in Hep3B cells in a dose-dependent manner. The strategy used here, by which a stable cell line with a low background and high fold Dox-induced expression of BNIPL-2 was generated, seemed to be more efficient and rapid for regulating BNIPL-2 gene expression.
The cDNA array technology was used to examine simultaneously the expression of specific genes on a single hybridization. Although expression analysis techniques such as differential display polymerase chain reaction (DD-PCR), Northern blot, serial analysis of gene expression (SAGE), and RT-PCR have been widely used, these studies are time-consuming and can only be used to deal with a limited number of genes. Thus a systematic approach to the examination of a large number of genes simultaneously is required. We therefore explored the gene expression profiles in human hepatocarcinoma cell line Hep3B regulated by BNIPL-2 overexpression using Atlas human cDNA array.
Following overexpression of BNIPL-2 in Hep3B cells, the expression level of 15 genes was altered, of which 8 were up-regulated and 7 were down-regulated. Genes involved in growth inhibition (IGFBP-1, TGF-β) or cell apoptosis (p21, E2A) were up-regulated. IGFBP-1 is thought to be the major short-term modulator of IGF bioavailability. In most clinical studies, circulating IGFBP-1 has been associated with growth inhibition[15]. Insulin-like growth factor I promotes muscle cell survival as an early event in differentiation through a pathway that involves Akt-dependent induction of p21[16]. Transforming growth factor-β (TGF-β) is a potent growth inhibitor for a wide variety of cells, controlling growth, differentiation and apoptosis of cells, and having important functions during embryonic development[17]. It has been found that Cdk inhibitor p21 is often responsible for stress-induced p53-dependent and p53-independent cell cycle arrest, and p21 plays an essential role in growth arrest after DNA damage and its overexpression leads to G1 and G2 or S-phase arrest. In addition, p21 is a negative regulator of p53-dependent apoptosis[18]. Human gastric cells were initially arrested in G1 by TGF-β and then driven to apoptosis after caspase-3-induced cleavage of p21[19]. Overexpression of E2A protein activity has been shown to induce either growth arrest or apoptosis, indicating that these proteins may regulate cell proliferation and survival[20,21]. Genes involved in cellular proliferation (p38 MAPK, Tob, CLK-1) were down-regulated in Hep3B cells when BNIPL-2 was overexpressed. P38 MAPK participates in not only inflammatory responses but also stress-induced signaling, cell proliferation and apoptosis. The p38 MAPK pathway participates in p21 induction, which consequently leads to decrease of cyclin D1/ cdk4 and cyclinE/cdk2 complexes[22]. Tob is a member of the Tob and BTG anti-proliferative protein family. Recently, one report indicated that Tob functioned as a molecular switch that regulates cell cycle progression through early G1[23]. In CLK-1 mutants, the affected features included cell cycle, embryonic and post-embryonic development, rhythmic behaviors, reproduction, and life span, all of which were lengthened or slowed down on average[24,25].
In conclusion, cDNA array is a powerful tool to explore gene expression profiles and the present results provide the basis for elucidating previous findings that BNIPL-2 might inhibit cell growth and promote apoptosis in BEL-7402 cells[2]. Although the mechanism of this interaction between BNIPL-2 and other proteins is not clear, we suggest that BNIPL-2 maybe inhibited growth by inducing cell cycle arrest at the G1 phase by TGF-β and p21 and affecting G2/M checkpoint through the p38 MAP kinase pathway.
Edited by Liu HX, Wang XL Proofread by Xu FM
| 1. | Shen L, Hu J, Lu H, Wu M, Qin W, Wan D, Li YY, Gu J. The apoptosis-associated protein BNIPL interacts with two cell proliferation-related proteins, MIF and GFER. FEBS Lett. 2003;540:86-90. |
| 2. | Qin W, Hu J, Guo M, Xu J, Li J, Yao G, Zhou X, Jiang H, Zhang P, Shen L. BNIPL-2, a novel homologue of BNIP-2, interacts with Bcl-2 and Cdc42GAP in apoptosis. Biochem Biophys Res Commun. 2003;308:379-385. |
| 3. | Zhou YT, Soh UJ, Shang X, Guy GR, Low BC. The BNIP-2 and Cdc42GAP homology/Sec14p-like domain of BNIP-Salpha is a novel apoptosis-inducing sequence. J Biol Chem. 2002;277:7483-7492. |
| 4. | Boyd JM, Malstrom S, Subramanian T, Venkatesh LK, Schaeper U, Elangovan B, D'Sa-Eipper C, Chinnadurai G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994;79:341-351. |
| 5. | Lottmann H, Vanselow J, Hessabi B, Walther R. The Tet-On system in transgenic mice: inhibition of the mouse pdx-1 gene activity by antisense RNA expression in pancreatic beta-cells. J Mol Med (Berl). 2001;79:321-328. |
| 6. | Milo-Landesman D, Surana M, Berkovich I, Compagni A, Christofori G, Fleischer N, Efrat S. Correction of hyperglycemia in diabetic mice transplanted with reversibly immortalized pancreatic beta cells controlled by the tet-on regulatory system. Cell Transplant. 2001;10:645-650. |
| 7. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. |
| 8. | Selaru FM, Zou T, Xu Y, Shustova V, Yin J, Mori Y, Sato F, Wang S, Olaru A, Shibata D. Global gene expression profiling in Barrett's esophagus and esophageal cancer: a comparative analysis using cDNA microarrays. Oncogene. 2002;21:475-478. |
| 9. | Rew DA. DNA microarray technology in cancer research. Eur J Surg Oncol. 2001;27:504-508. |
| 10. | Wang JR, Qin WX, Shu HQ, Pan Y, Zhang YY, Wan DF. Establishment of Hep3B cell line controlled by the Tet-On regulatory system. Tumor. 2002;22:191-193. |
| 11. | Sonntag KC, Haller GW, Giauffret D, Germana S, Reeves SA, Levy J, Sachs DH, LeGuern C. Regulated expression of an MHC class II gene from a promoter-inducible retrovirus. Hum Gene Ther. 2000;11:1961-1969. |
| 12. | Kashima Y, Miki T, Minami K, Seino S. Establishment of a tet-on gene expression system in glucose-responsive and -unresponsive MIN6 cells. Diabetes. 2001;50 Suppl 1:S133. |
| 13. | Wang D, Grammer JR, Cobbs CS, Stewart JE, Liu Z, Rhoden R, Hecker TP, Ding Q, Gladson CL. p125 focal adhesion kinase promotes malignant astrocytoma cell proliferation in vivo. J Cell Sci. 2000;113 Pt 23:4221-4230. |
| 14. | Kobayashi T, Sawa H, Morikawa J, Zhang W, Shiku H. Bax induction activates apoptotic cascade via mitochondrial cytochrome c release and Bax overexpression enhances apoptosis induced by chemotherapeutic agents in DLD-1 colon cancer cells. Jpn J Cancer Res. 2000;91:1264-1268. |
| 15. | Kajantie E, Hytinantti T, Koistinen R, Risteli J, Rutanen EM, Seppälä M, Andersson S. Markers of type I and type III collagen turnover, insulin-like growth factors, and their binding proteins in cord plasma of small premature infants: relationships with fetal growth, gestational age, preeclampsia, and antenatal glucocorticoid treatment. Pediatr Res. 2001;49:481-489. |
| 16. | Lawlor MA, Rotwein P. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol Cell Biol. 2000;20:8983-8995. |
| 17. | Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117-129. |
| 18. | Zhang Y, Fujita N, Tsuruo T. Caspase-mediated cleavage of p21Waf1/Cip1 converts cancer cells from growth arrest to undergoing apoptosis. Oncogene. 1999;18:1131-1138. |
| 19. | Kim DK, Cho ES, Lee SJ, Um HD. Constitutive hyperexpression of p21(WAF1) in human U266 myeloma cells blocks the lethal signaling induced by oxidative stress but not by Fas. Biochem Biophys Res Commun. 2001;289:34-38. |
| 20. | Park ST, Nolan GP, Sun XH. Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. J Exp Med. 1999;189:501-508. |
| 21. | Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc Natl Acad Sci USA. 1999;96:996-1001. |
| 22. | Moon SK, Jung SY, Choi YH, Lee YC, Patterson C, Kim CH. PDTC, metal chelating compound, induces G1 phase cell cycle arrest in vascular smooth muscle cells through inducing p21Cip1 expression: involvement of p38 mitogen activated protein kinase. J Cell Physiol. 2004;198:310-323. |
| 23. | Suzuki T, K-Tsuzuku J, Ajima R, Nakamura T, Yoshida Y, Yamamoto T. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 2002;16:1356-1370. |
| 25. | Levavasseur F, Miyadera H, Sirois J, Tremblay ML, Kita K, Shoubridge E, Hekimi S. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J Biol Chem. 2001;276:46160-46164. |