Published online Apr 1, 2004. doi: 10.3748/wjg.v10.i7.1069
Revised: November 23, 2003
Accepted: December 16, 2003
Published online: April 1, 2004
AIM: An insertion mutation at nucleotide 3020 (3020insC) in the Caspase recruitment domain gene (CARD15), originally reported as NOD2, is strongly associated with Crohn’s disease. The C-insertion mutation at nucleotide 3020 (3020inC) in the leucine-rich repeat (LRR) region results in a frameshift in the 10th LRR followed by a premature stop codon. This truncation mutation is responsible for the inability to activate nuclear factor (NF)-κB in response to bacterial lipopolysaccharide (LPS). The present study aimed to genotype NOD2/CARD15 gene 3020insC frameshift mutation in Chinese patients with inflammatory bowel disease.
METHODS: We genotyped an insertion polymorphism affecting the leucine-rich region of the protein product by the allele specific PCR in 74 unrelated patients with ulcerative colitis of Han nationality in Hubei Province of China, 15 patients with Crohn’s disease and 172 healthy individuals.
RESULTS: No significant differences were found in the genotype and allele frequencies of the C-insertion mutation of NOD2 gene among patients with Crohn’s disease and ulcerative colitis and healthy controls.
CONCLUSION: NOD2 gene 3020insC frameshift mutation is not a major contributor to the susceptibility to both Crohn’s disease and ulcerative colitis in Chinese Han patients.
- Citation: Guo QS, Xia B, Jiang Y, Qü Y, Li J. NOD2 3020insC frameshift mutation is not associated with inflammatory bowel disease in Chinese patients of Han nationality. World J Gastroenterol 2004; 10(7): 1069-1071
- URL: https://www.wjgnet.com/1007-9327/full/v10/i7/1069.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i7.1069
Inflammatory bowel disease (IBD) is a chronic intestinal inflammatory disorder that is clinically classified into Crohn’s disease (CD), ulcerative colitis (UC) and indeterminate colitis. IBD is characterized by a dysregulated mucosal immune response and its pathogenesis has not been fully illustrated, but several epidemiological and genetic studies have suggested that IBD is predisposed by certain environmental and genetic factors[1-3]. IBD1 was the first susceptible locus linked to Crohn’s disease. Utilizing genome-wide linkage studies among families with multiple affected members, Hugot et al[1] mapped the IBD1 gene to the proximal region of the long arm of chromosome 16 (16q12) in the white population. In Jewish families, the IBD1 locus was demonstrated to be significantly associated with CD[4], but not with ulcerative colitis. Hugot et al[5] identified nucleotide oligomerisation domain (NOD2) as the IBD1 gene through routine positional cloning methods, the same finding was also reported in several other studies[6,7]. NOD2, a member of the NOD1/APAF1 gene family, comprises an amino-terminal effector domain, a nucleotide-binding domain and leucine-rich repeats (LRRs)[8]. Recently, NOD2 has been known as Caspase activating recruitment domain (CARD) 15. NOD2/CARD molecule is expressed exclusively in monocytes and activates NF-κB through the interaction with its N-terminal CARDs[8]. As studies have shown that the signaling by tumor necrosis factor (TNF) and activation by nuclear factor (NF)-κB play a key role in IBD, NOD2 mutation may lead to the variation of NF-κB activation. Three mutations (R702W, G908R, 1007fs, respectively at SNP8, 12, and 13) have been identified in NOD2/CARD15, which were shown to be independently associated with CD. One of the mutations predisposing to CD is 1007fs, the C-insertion mutation at nucleotide 3020 (3020inC) in the leucine-rich repeat region, results in a frameshift in the 10th LRR followed by a premature stop codon. This truncation mutation may result in the inability to activate NF-κB in response to bacterial lipopolysaccharides (LPS). Therefore, NOD2 is considered to play a role in the pathogenesis of CD.
Studies by Japanese and Hongkong researchers, however, have failed to identify the insertion mutation both in patients and in control subjects[9,10]. Greek has shown that NOD2 is not significant in the pathogenesis of CD[11]. These studies support that CD possesses genetic heterogeneity among different populations. In this present study, we genotyped the 3020insC mutation of NOD2 gene in CD and UC in Chinese subjects of Han nationality in Hubei Province, to identify the susceptible gene for Chinese.
A total of 15 unrelated patients with CD and 74 patients with UC were included in this study. As a control group 174 healthy individuals were also recruited from medical staff and students in Zhongnan Hospital of Wuhan University, as well as from healthy volunteers in Wuhan city. The diagnosis of either UC or CD was established and verified by clinical, radiological, endoscopic and histological examinations in accordance with the criteria by Lennard-Johns[12]. All the subjects included in this study were unrelated Chinese of Han nationality in Hubei Province. This study protocol was approved by the Ethic Committee of Medical School of Wuhan University.
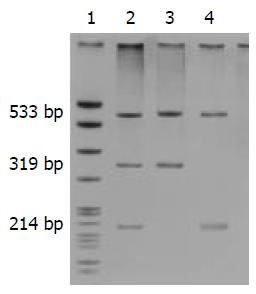
DNA was isolated from peripheral blood leukocytes by conventional proteinase K digestion and phenol/chloroform extraction methods. Allele-specific PCR was employed to detect the 3020insC mutation of NOD2 gene[6] and to generate a nonspecific 533-bp product, along with a 319-bp fragment (wild type) and/or a 214-bp fragment (3020inC). The primers used for PCR are listed in Table 1. PCR was performed in a reaction system in a volume of 25 μL containing sense and antisense primers (each in a volume of 0.5 μL at the concentration of 20 pmol/μL ), wild-type sense and 3020insC antisense primer respectively (each in a volume of 0.25 μL at the concentration of 20 pmol/μL), 2.5 μL of 10×buffer, 0.5 μL of 10 mmol/L dNTPs, 1 μL of template DNA, 0.5 μL of AmpliTaq DNA polymerase (MBI Fermentas), and 19 μL of ddH2O. Amplification was carried out with a Perkin-Elmer thermocycler. The cycling was performed with an initial denaturation for 5 min at 94 °C, followed by 35 cycles at 94 °C for 45 s, at 59 °C for 45 s, at 72 °C for 45 s with a final extension at 72 °C for 7 min. The PCR products were identified by electrophoresis in non-denaturing polyacrylamide gels containing 8% acrylamide-bisacrylamide (29:1), 0.5 × Tris-borate-EDTA (TBE), 100 g/L ammonium persulfate, and TEMED at 150 V for 1.5 h. The gels were then subjected to silver staining. The PCR products used for sequencing analyses were purified and analyzed using an ABI377 automated sequencer (Applied Biosystems, USA). DNA samples for genotype control analysis were kindly provided by the Laboratory of Immunogenetics of Free University of Amsterdam, Netherlands.
| Primer | Sequence |
| Sense primer | 5’-CTGAGCCTTTGTTGATGAGC-3’ |
| Antisense primer | 5’-TCTTCAACCACATCCCCATT-3’ |
| Wild-type sense primer | 5’-CAGAAGCCCTCCTGCAGGCCCT-3’ |
| 3020insC antisense primer | 5’-CGCGTGTCATTCCTTTCATGGGGC-3’ |
Statistical analysis was performed with SPSS 11.5 software package. The data were analyzed by χ2 test or Fisher’s exact test. A P value less than 0.05 was considered statistically significant. The confidential intervals (CI) for the odds ratios (OR) were calculated by Woolf formula.
The results of the genotypes and allele frequencies of the 3020insC mutation in CD and UC patients and in ethnically matched Chinese healthy controls are shown in Table 2. Two heterozygotes of the mutation in UC patients and one in CD patients were identified, whereas only one heterozygote mutation was found in healthy controls. No significant associations were noted in genotypes and allele frequencies of the 3020insC mutation in UC (P = 0.2165) or CD (P = 0.1542) in comparison with the healthy controls. The NOD2 3020insC mutation was not associated with CD or UC in Hubei Han population (Figure 1).
| Genome | Healthycontrols(n=172) | Ulcerativecolitis(n=74) | Crohn’sdisease(n=15) |
| Wild type | 171 | 72 | 14 |
| Heterozygote | 1 | 2 | 1 |
| 3020insC mutation homozygote | 0 | 0 | 0 |
| Wild type allele frequency | 99.7% | 98.7% | 96.7% |
| Mutative allele frequency | 0.3% | 1.3% | 3.3% |
In this study, we did not find any evidence suggestive of the association of NOD2 frameshift mutations with CD or UC. The allele frequency of 3020insC mutation was slightly increased in CD and UC patients when compared with the healthy controls, but there were no statistically significant differences among these three groups. After the initial report of the associations between mutations in the NOD2 gene and susceptibility to CD[5,6], a German and British study[7] soon found evidence to show that the insertion mutation in the NOD2 gene was responsible for a substantially increased susceptibility to CD but not to UC. Other studies have suggested that attributive risks of the 3020insC mutation of NOD2 differ in different ethnic nationalities and countries. A large-scale study in Japanese failed to identify the insertion mutation both in patients and in controls[9], the same result was also reported by Leong et al[10] in Chinese population in Hong Kong. A study in a Cretan showed that NOD2/CARD15 was not significant in the pathogenesis of CD[11]. All these data and the results of this study demonstrate that CD has genetic heterogeneity among different nationalities.
NOD2 was recently identified as a susceptible gene of CD in most Western countries, which provided an opportunity to study the relationship between NOD2 gene and innate immunity in CD. NOD2 was located in the peak region of linkage on chromosome 16q12 and a member of the NOD1/APAF1 gene family[8], expressed primarily in monocytes and functioned to activate NF-κB in response to LPS stimulation[13,14]. 3020insC, which resulted in a frameshift mutation at the second nucleotide of codon 1007, a Leu1007→Pro substitution in the tenth LRR and followed by a premature stop codon, encoded a truncated NOD2 protein and led to a weakened innate immune response in CD[8]. In our study we did not find any statistically significant association between the 3020insC mutation and CD. Other mechanisms of CD involving the susceptible gene and innate immunity have been suggested in Chinese Han nationality in Hubei Province.
Edited by Chen WW and Wang XL Proofread by Xu FM
| 1. | Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 598] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 2. | Cavanaugh J. International collaboration provides convincing linkage replication in complex disease through analysis of a large pooled data set: Crohn disease and chromosome 16. Am J Hum Genet. 2001;68:1165-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 149] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 3. | Curran ME, Lau KF, Hampe J, Schreiber S, Bridger S, Macpherson AJ, Cardon LR, Sakul H, Harris TJ, Stokkers P. Genetic analysis of inflammatory bowel disease in a large European cohort supports linkage to chromosomes 12 and 16. Gastroenterology. 1998;115:1066-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 110] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 4. | Akolkar PN, Gulwani-Akolkar B, Lin XY, Zhou Z, Daly M, Katz S, Levine J, Present D, Gelb B, Desnick R. The IBD1 locus for susceptibility to Crohn's disease has a greater impact in Ashkenazi Jews with early onset disease. Am J Gastroenterol. 2001;96:1127-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3901] [Article Influence: 162.5] [Reference Citation Analysis (0)] |
| 6. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3555] [Cited by in RCA: 3474] [Article Influence: 144.8] [Reference Citation Analysis (1)] |
| 7. | Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925-1928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 769] [Cited by in RCA: 769] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 8. | Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812-4818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1040] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 9. | Yamazaki K, Takazoe M, Tanaka T, Kazumori T, Nakamura Y. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn's disease. J Hum Genet. 2002;47:469-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 201] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 10. | Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, Sung JJ. NOD2/CARD15 gene polymorphisms and Crohn's disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 11. | Roussomoustakaki M, Koutroubakis I, Vardas EM, Dimoulios P, Kouroumalis EA, Baritaki S, Koutsoudakis G, Krambovitis E. NOD2 insertion mutation in a Cretan Crohn's disease population. Gastroenterology. 2003;124:272-23; author reply 272-23;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2-6; discussion 16-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1396] [Cited by in RCA: 1445] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 13. | Neurath MF, Pettersson S, Meyer zum Buschenfelde KH, Strober W. Local administration of antisense phosphorothioate oligonucleotides to the p65 subunit of NF-kappa B abrogates established experimental colitis in mice. Nat Med. 1996;2:998-1004. [RCA] [DOI] [Full Text] [Cited by in Crossref: 618] [Cited by in RCA: 631] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 14. | Schreiber S, Nikolaus S, Hampe J. Activation of nuclear factor kappa B inflammatory bowel disease. Gut. 1998;42:477-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 580] [Article Influence: 21.5] [Reference Citation Analysis (0)] |