Published online Feb 15, 2004. doi: 10.3748/wjg.v10.i4.531
Revised: July 12, 2003
Accepted: July 24, 2003
Published online: February 15, 2004
AIM: To develop a novel non-viral gene delivery system, which has a small particle size and a high transfection efficiency to hepatocyte and hepatoma cells.
METHODS: Lipid-polycation-DNA lipopolyplex (LPD) was prepared by mixing plasmid DNA and polylysine. The resulted polyplex was incubated for 10 min at room temperature, following the addition of preformed cationic liposomes. The morphology of LPD was observed by transmission electron microscopy. The diameter and surface charge of LPD were measured by photon correlation spectroscopy (PCS). The nuclease protection ability of LPD was evaluated by agarose gel electrophoresis. Estimation of the transfection efficiency was performed by galactosidase assay in Chang cells and SMMC-7721 cells.
RESULTS: LPD had a regular spherical surface. The average diameter and the zeta potential of LPD were 132.1 nm and 26.8 mV respectively. LPD could protect plasmid DNA from nuclease degradation after 2 hours incubation at 37 °C while the naked DNA degraded rapidly. The average transfection efficiencies were 86.2% ± 8.9% and 72.4% ± 6.5% in Chang cells and SMMC-7721 cells respectively.
CONCLUSION: LPD has a rather small particle size and a high transfection activity. LPD may be a good non-viral vector for application in some gene delivery.
- Citation: Sun X, Zhang HW, Zhang ZR. Transfection efficiency of pORF lacZ plasmid lipopolyplex to hepatocytes and hepatoma cells. World J Gastroenterol 2004; 10(4): 531-534
- URL: https://www.wjgnet.com/1007-9327/full/v10/i4/531.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i4.531
Gene therapy focuses on the therapeutic use of genes, and has achieved considerable advances in the treatment of both acquired and inherited diseases[1,2]. The success of gene therapy rests on the development of a vector that can selectively and efficiently deliver a gene to the target cells with minimal toxicity[3,4]. The vectors used to date can be classified into viral and non-viral groups.
Non-viral delivery systems for gene therapy have been increasingly proposed as safer alternatives to viral vectors. They have the potential to be repeatedly used with minimal host immune response and are targetable, stable in storage, and easy to produce in large quantities. These advantages have provided the impetus to continue their development. So far, several non-viral delivery systems have been developed, such as liposomes[5-7], nanoparticles[8-10] hydrogel[11] emulsion[12] and peptide nucleic acid[13]. Complexes formed between cationic liposomes and plasmid DNA are the predominant non-viral vectors employed for the transfection of eukaryotic cells in research laboratories[14-16]. Currently, several cationic liposomal formulations have also undergone clinical evaluation as vectors for gene therapy in cancer and cystic fibrosis. However, the efficiency and specificity of non-viral delivery systems are not so high. To improve the transfection efficiency, some cationic lipids[17] and polymers[18-20] have been synthesized. Ligand or antibody mediated targeting of gene transfer has also been widely explored[21-24]. Furthermore, nuclear localization sequences (NLS) are studied to help the entry of plasmid DNA from cytoplasm into nucleus[25].
The liver possesses a variety of characteristics that make this organ very attractive for gene therapy[26,27]. The proportion of administered macromolecules internalized by hepatocytes depends on their particle size and biochemical characteristics. Only relatively small molecules can pass the fenestrate of sinusoidal endothelial cells of the liver, since their diameter is about 100 nm[28,29]. Polycations as a formulation component have been shown to enhance the efficiency of liposomes-mediated gene transfer both in vitro and in vivo. Specifically, lipid-polycation-DNA lipopolyplexes (LPD) have appeared promising as efficient gene-delivery vehicles for systemic administration[30-33]. In this study, we developed a novel lipopolyplex formulation, which is small in particle size and high in transfection efficiency to hepatocytes and hepatoma cells.
Plasmid pORF lacZ (3.54 kb) was purchased from Invivogen (USA). Poly-L-lysine (PLL, Mr29000), dimethyldioc tadecyl ammonium bromide (DDAB) and β-galactosidase reporter gene staining kit were purchased from Sigma. Hepatoma cell line SMMC-7721 and hepatocyte cell line Chang were obtained from Shanghai Cell Institute, China Academy of Sciences. Cell culture media DMEM and RPMI 1640 were obtained from Gibco Co. (USA). Qiagen Giga Endo-free plasmid purification kit was purchased from Qiagen (CA.USA). All the other chemicals and reagents used were of the analytical grade obtained commercially.
Plasmid pORF lacZ (3.54 kb), is a eukaryotic expression vector containing the EF-1α/HTLV hybrid promoter within an intron. The lacZ gene codes for the enzyme β-galactosidase, whose activity allows for quick determination of cells expressing the lacZ gene. pORF-lacZ plasmid DNA was isolated and purified from DH5-α E coli using the Qiagen Giga Endo-free plasmid purification kit. DNA concentration and purity were quantified by UV absorbance at 260 nm and 280 nm on a GBC UV cintra 10e spectrophotometer. The structural integrity and topology of purified DNA were analyzed by agarose gel electrophoresis.
Cationic liposomes composed of DDAB/cholesterol were prepared with the molar ratio of 1:1. The lipid mixture was dissolved in appropriate chloroform and a thin lipid film was formed in a round-bottomed flask by drying the solvent using a rotary evaporator. The film was hydrated at 60 °C with the addition of 10 mM herpes buffer (pH7.4). The lipids were resuspended and then undergone ten passes through an extruder with 200, 100 nm polycarbonate membranes respectively. LPDs were formed by mixing equal volumes of DNA and PLL. DNA and PLL were both diluted from the stock with 10 Mm herpes buffer. After mixed, the solution was briefly vortexed, and the resulting polyplexes were incubated for 10 min at room temperature. Concentrated cationic liposomes were subsequently added to the DNA/PLL mixture to achieve the desired final component concentrations and ratios.
Diameter and surface charge of lipopolyplexes were measured by photon correlation spectroscopy (PCS) (Malvern zetasizer 3000 HS, Malvern Instruments Ltd., UK) with a 50 mV laser. Twenty µl of LPD was diluted by 3 ml of 10 mM herpes buffer and added into the sample cell. The measurement time was set at 2 min (rapid measurement) and each run consisted of 10 subruns[34]. The measurements were done at 25 °C at an angel of 90°. The size distribution followed a lognormal distribution. The potential of the lipid carriers at the surface of spheres, called the zeta potential(ξ), was derived from the mobile particles in electric field by applying the smoluchowsky relationship, which was measured at least three times and at an average of appropriate concentrations of samples.
Just prior to use, the former-coated 100-mesh copper girds were prepared by glow discharging. The girds were then floated on 25 µl of samples for 90 s, wicked off, and floated on drops of 1% aqueous uranyl acetate for 90 s. Finally, the samples were wicked off, dried in air and stored at room temperature. Negative stain electron micrographs of LPD were taken using a JEM-100SX electron microscope.
Naked DNA or LPDs were incubated with DNase I solution (0.32 U/µg DNA) at 37 °C for 5 min, 1 h and 2 h respectively. The enzyme reaction was stopped by addition of 0.5M EDTA[35]. Triton X-100 (final concentration 1% v/v) was added to destroy the bilayer structure of liposomes and 0.9% w/v heparin was added to release DNA from PLL/DNA complexes[36]. The samples were carefully added to the wells of a 0.8% agarose gel at a volume (representing 1 µg of DNA per well). The gel was run in TBE buffer containing 0.5 µg/ml EtBr at 100V for 1 h. Subsequently, the gel was removed from the tank and visualized under UV light by molecular analyst software.
Chang cells and SMMC-7721 cells were cultured in DMEM and RPMI-1640 respectively with 10% fetal bovine serum and streptomycin (100 µg/ml). The cells were seeded at 2 × 105 cells per well onto 6-well plates 24 h before transfection. The cells were about 70% confluence at the time of transfection. Then the cells were washed twice by PBS, and 1 ml of serum-free and antibiotics-free medium was added into each well[37]. For each well in a transfection, LPDs containing 5 µg pORF-1acZ were overlaid and mixed gently. The cells were incubated with LPD for 5 hours at 37 °C in a CO2 incubator. Following incubation, LPD was removed and the cell surfaces were rinsed thoroughly and treated with 2 ml fresh complete medium. Then the cells were returned to the incubator for a further 45 h to allow intracellular gene expression to proceed.
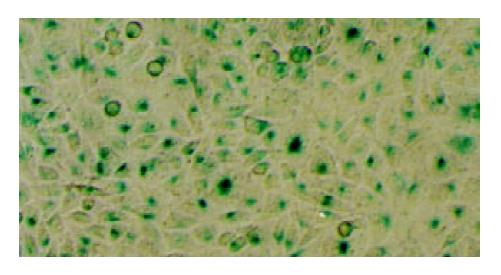
Estimation of the transfection efficiency was performed using galactosidase assay[38,39]. After the desired time of incubation, the cells were washed with PBS twice and fixed with 2% formaldehyde and 0.2% glutaraldehyde for 10 minutes at room temperature. Then the cells were rinsed twice and stained by X-gal (20 mg/ml) according to the manufacture’s instructions. The cells were incubated at 37 °C overnight and observed under a microscope. The transfected cells were blue after X-gal staining. For each well, five visual fields were chosen randomly. Cells stained blue were counted and the transfection efficiency was calculated as the percentage of the blue cells in each field.
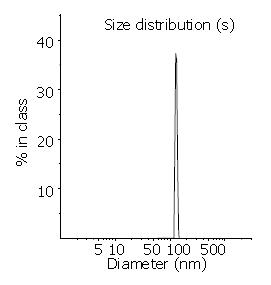
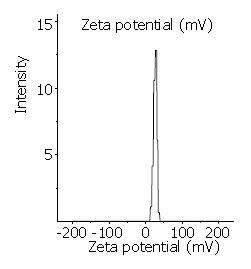
Transmission electron microscopy (Figure 1) demonstrated the regular spherical surface of LPD. Figure 2 shows the average diameter of LPD being 132.1 nm with a very narrow distribution (polyindex 0.148). Figure 3 illustrates the zeta potential of LPD being 26.8 mV in 10 mM herpes buffer (pH7.4).
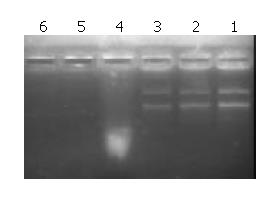
Figure 4 shows that LPD could protect plasmid DNA from nuclease degradation after 5 minutes, 1 hour and 2 hours of incubation at 37 °C while the naked DNA degraded rapidly.
Figure 5 and Figure 6 demonstrate that LPD had a rather high transfection efficiency both in Chang cells and in SMMC-7721 cells. The average transfection efficiencies were 86.2% ± 8.9% and 72.4% ± 6.5% in Chang cells and SMMC-7721 cells, respectively.
The amount of intact DNA present in LPD cannot be directly assayed by gel electrophoresis as the complexation hinders the binding between DNA and ethidium bromide. Therefore Triton X-100, a widely used detergent is employed to destroy the liposomal bilayer. Furthermore, dissociation of cationic polymer and DNA is also required. Traditionally, DNA is dissociated from PLL by digestion with trypsin and phenol extraction. This procedure is time-consuming and the lost DNA during extraction is immeasurable. Some new dissociation methods have been established based on the fact that the interaction between DNA and PLL was mainly due to electrostatic bonds[40-43]. By raising the pHof electrophoretic buffer above the pKa of PLL, PLL could become less protonated, thus reducing the charge and thereby allowing the complexes to disossociation under electrophoretic conditions. However, when pHwas higher than 11.6, ethidium bromide would lose its affinity for DNA and fluorescence was lost. In this study, heparin at final concentration of 0.9% (w/v) was added to release DNA from the complexes[36]. Heparin, as an anionic polysaccharide can bind to PLL by electrostatic interaction. When enough amount of heparin was added, DNA could be completely released from PLL-DNA complexes. The presence of heparin did not interfere with the gel electrophoresis, so the sample could be loaded directly.
The size and surface charge of lipopolyplex could influence their physical stability, in vivo distribution, cellular interaction and extent of cell uptake. After intravenous administration, particles larger than 7 µm are normally filtered by the smallest capillaries of the lungs, and particles smaller than 7 µm in diameter may pass the smallest lung capillary beds and be entrapped in the capillary network of the liver and spleen. Particles between 100 nm and 2 µm in size are rapidly cleared from the bloodstream by the mononuclear phagocytic system (MPS). Typically in practice, 80% - 90% of hydrophobic particles were opsonized and taken up by fixed macrophages of the liver and spleen, often within a few minutes of intravenous administration[1]. Zeta potential measurements can be a useful tool for characterizing colloidal drug delivery systems. They can give information about the surface properties of the carrier and therefore helping determine how the constituent molecules are organized. In this study, the LPDs had a positive zeta potential of 26.8 mV. Therefore, they could interact with negatively charged cell surface, which resulted in cellular internalization. Furthermore, plasmid DNA in LPDs was shielded from nuclease digestion due to the formation of charge complexes.
It has become clear that cationic liposome plays several roles in the process of transfection, such as condensing and protecting DNA, binding to cell surface, triggering endocytosis and releasing DNA/lipid complexes from endosome. The size of condensed DNA is thought to be critical for in vivo delivery because the particle size influences not only the biodistribution but also the efficiency of cellular uptake through endocytosis. At an appropriate condition, cationic polymer PLL can precondense plasmid DNA more effectively than cationic liposomes. Therefore, LPD has a rather small particle size and a high transfection activity in hepatocytes and hepatoma cells. LPD may be a superior vector for some applications in gene delivery compared to regular DNA/liposome complexes. Structurally, this formulation has been found to be a virus-like particle, each containing a condensed genome as the core and a lipidic shell as the envelope. The liver possesses a variety of characteristics which make this organ very attractive for gene therapy. Although some of the virus-mediated gene transfer systems have been found to be quite effective, their usefulness is limited, given that they induce an immune response, leading to the rapid rejection of transduced cells. To overcome this problem, our further work will focus on the use of LPD in the treatment of liver cancer.
Edited by Wu XN and Wang XL
| 1. | Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46:187-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 237] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 2. | Liu F, Huang L. Development of non-viral vectors for systemic gene delivery. J Control Release. 2002;78:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 196] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 3. | Nishikawa M, Huang L. Nonviral vectors in the new millennium: delivery barriers in gene transfer. Hum Gene Ther. 2001;12:861-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 405] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 4. | Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13:128-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 301] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 5. | Stuart DD, Allen TM. A new liposomal formulation for antisense oligodeoxynucleotides with small size, high incorporation efficiency and good stability. Biochim Biophys Acta. 2000;1463:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Bailey AL, Sullivan SM. Efficient encapsulation of DNA plasmids in small neutral liposomes induced by ethanol and calcium. Biochim Biophys Acta. 2000;1468:239-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Sudimack JJ, Guo W, Tjarks W, Lee RJ. A novel pH-sensitive liposome formulation containing oleyl alcohol. Biochim Biophys Acta. 2002;1564:31-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 8. | Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55:329-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2387] [Cited by in RCA: 2034] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 9. | Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, August JT, Leong KW. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 824] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 10. | Corsi K, Chellat F, Yahia L, Fernandes JC. Mesenchymal stem cells, MG63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials. 2003;24:1255-1264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 260] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 11. | Vinogradov SV, Bronich TK, Kabanov AV. Nanosized cationic hydrogels for drug delivery: preparation, properties and interactions with cells. Adv Drug Deliv Rev. 2002;54:135-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 504] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 12. | Kim YJ, Kim TW, Chung H, Kwon IC, Sung HC, Jeong SY. The effects of serum on the stability and the transfection activity of the cationic lipid emulsion with various oils. Int J Pharm. 2003;252:241-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA). Adv Drug Deliv Rev. 2003;55:267-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 206] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 14. | Audouy SA, de Leij LF, Hoekstra D, Molema G. In vivo characteristics of cationic liposomes as delivery vectors for gene therapy. Pharm Res. 2002;19:1599-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 15. | Lesage D, Cao A, Briane D, Lievre N, Coudert R, Raphael M, Salzmann Jl, Taillandier E. Evaluation and optimization of DNA delivery into gliosarcoma 9L cells by a cholesterol-based cationic liposome. Biochim Biophys Acta. 2002;1564:393-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Templeton NS. Cationic liposome-mediated gene delivery in vivo. Biosci Rep. 2002;22:283-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Ewert K, Ahmad A, Evans HM, Schmidt HW, Safinya CR. Efficient synthesis and cell-transfection properties of a new multivalent cationic lipid for nonviral gene delivery. J Med Chem. 2002;45:5023-5029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 117] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 18. | Kabanov AV, Lemieux P, Vinogradov S, Alakhov V. Pluronic block copolymers: novel functional molecules for gene therapy. Adv Drug Deliv Rev. 2002;54:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 254] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Maheshwari A, Mahato RI, McGregor J, Han So, Samlowski WE, Park JS, Kim SW. Soluble biodegradable polymer-based cytokine gene delivery for cancer treatment. Mol Ther. 2000;2:121-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 20. | Lim YB, Han SO, Kong HU, Lee Y, Park JS, Jeong B, Kim SW. Biodegradable polyester, poly[α-(4-aminobutyl)-L-glycolic acid], as a non-toxic gene carrier. Pharm Res. 2000;17:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 123] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Li WM, Mayer LD, Bally MB. Prevention of antibody-mediated elimination of ligand-targeted liposomes by using poly(ethylene glycol)-modified lipids. J Pharmacol Exp Ther. 2002;300:976-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Mastrobattista E, Kapel RH, Eggenhuisen MH, Roholl PJ, Crommelin DJ, Hennink WE, Storm G. Lipid-coated polyplexes for targeted gene delivery to ovarian carcinoma cells. Cancer Gene Ther. 2001;8:405-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Arangoa MA, Düzgüneş N, Tros de Ilarduya C. Increased receptor-mediated gene delivery to the liver by protamine-enhanced-asialofetuin-lipoplexes. Gene Ther. 2003;10:5-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Fisher KD, Ulbrich K, Subr V, Ward CM, Mautner V, Blakey D, Seymour LW. A versatile system for receptor-mediated gene delivery permits increased entry of DNA into target cells, enhanced delivery to the nucleus and elevated rates of transgene expression. Gene Ther. 2000;7:1337-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 82] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Escriou V, Carrière M, Scherman D, Wils P. NLS bioconjugates for targeting therapeutic genes to the nucleus. Adv Drug Deliv Rev. 2003;55:295-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 119] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Ghosh SS, Takahashi M, Thummala NR, Parashar B, Chowdhury NR, Chowdhury JR. Liver-directed gene therapy: promises, problems and prospects at the turn of the century. J Hepatol. 2000;32:238-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Hwang SH, Hayashi K, Takayama K, Maitani Y. Liver-targeted gene transfer into a human hepatoblastoma cell line and in vivo by sterylglucoside-containing cationic liposomes. Gene Ther. 2001;8:1276-1280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Jain RK. Delivery of molecular and cellular medicine to solid tumors. Adv Drug Deliv Rev. 2001;46:149-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 401] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 29. | Christie RJ, Grainger DW. Design strategies to improve soluble macromolecular delivery constructs. Adv Drug Deliv Rev. 2003;55:421-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Guo W, Gosselin MA, Lee RJ. Characterization of a novel diolein-based LPDII vector for gene delivery. J Control Release. 2002;83:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Birchall JC, Kellaway IW, Gumbleton M. Physical stability and in-vitro gene expression efficiency of nebulised lipid-peptide-DNA complexes. Int J Pharm. 2000;197:221-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Li B, Li S, Tan Y, Stolz DB, Watkins SC, Block LH, Huang L. Lyophilization of cationic lipid-protamine-DNA (LPD) complexes. J Pharm Sci. 2000;89:355-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 33. | Tsai JT, Furstoss KJ, Michnick T, Sloane DL, Paul RW. Quantitative physical characterization of lipid-polycation-DNA lipopolyplexes. Biotechnol Appl Biochem. 2002;36:13-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 34. | Dekie L, Toncheva V, Dubruel P, Schacht EH, Barrett L, Seymour LW. Poly-L-glutamic acid derivatives as vectors for gene therapy. J Control Release. 2000;65:187-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 101] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Cui Z, Mumper RJ. Plasmid DNA-entrapped nanoparticles engineered from microemulsion precursors: in vitro and in vivo evaluation. Bioconjug Chem. 2002;13:1319-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Moret I, Esteban Peris J, Guillem VM, Benet M, Revert F, Dasí F, Crespo A, Aliño SF. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76:169-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 37. | Dokka S, Toledo D, Shi X, Ye J, Rojanasakul Y. High-efficiency gene transfection of macrophages by lipoplexes. Int J Pharm. 2000;206:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 38. | Sakurai F, Nishioka T, Saito H, Baba T, Okuda A, Matsumoto O, Taga T, Yamashita F, Takakura Y, Hashida M. Interaction between DNA-cationic liposome complexes and erythrocytes is an important factor in systemic gene transfer via the intravenous route in mice: the role of the neutral helper lipid. Gene Ther. 2001;8:677-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 126] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 39. | Armeanu S, Pelisek J, Krausz E, Fuchs A, Groth D, Curth R, Keil O, Quilici J, Rolland PH, Reszka R. Optimization of nonviral gene transfer of vascular smooth muscle cells in vitro and in vivo. Mol Ther. 2000;1:366-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 40. | Männistö M, Vanderkerken S, Toncheva V, Elomaa M, Ruponen M, Schacht E, Urtti A. Structure-activity relationships of poly(L-lysines): effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J Control Release. 2002;83:169-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 41. | Parker AL, Oupicky D, Dash PR, Seymour LW. Methodologies for monitoring nanoparticle formation by self-assembly of DNA with poly(l-lysine). Anal Biochem. 2002;302:75-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 42. | Hill IR, Garnett MC, Bignotti F, Davis SS. Determination of protection from serum nuclease activity by DNA-polyelectrolyte complexes using an electrophoretic method. Anal Biochem. 2001;291:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 43. | Safinya CR. Structures of lipid-DNA complexes: supramolecular assembly and gene delivery. Curr Opin Struct Biol. 2001;11:440-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 291] [Article Influence: 12.1] [Reference Citation Analysis (0)] |