Published online Oct 15, 2004. doi: 10.3748/wjg.v10.i20.3006
Revised: November 26, 2003
Accepted: December 8, 2003
Published online: October 15, 2004
AIM: To analyze the tissue morphologic phenotype and liver gene expression profile of hB1F transgenic mice.
METHODS: Transgene expression was analyzed with RT-PCR and Western blotting. For one of the transgenic mouse lines, tissue expression pattern of the transgene was also examined with immunochemical methods. Pathological analysis was used to examine the tissue morphologic phenotype of established transgenic mice. The liver gene expression profile of transgenic mice was analyzed with microchip, and some of the differentially expressed genes were verified with RT-PCR.
RESULTS: The expressions of hB1F were shown in livers from 6 of 7 transgenic mouse lines. The overexpression of hB1F transgene did not cause pathological changes. Expressions of three genes were up-regulated, while down-regulation was observed for 25 genes.
CONCLUSION: The overexpression of hB1F transgene may cause changes of gene expression profiles in the liver of transgenic mice.
- Citation: Wang SL, Yang H, Xie YH, Wang Y, Li JZ, Wang L, Wang ZG, Fu JL. Gene expression profile in liver of hB1F transgenic mice. World J Gastroenterol 2004; 10(20): 3006-3010
- URL: https://www.wjgnet.com/1007-9327/full/v10/i20/3006.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i20.3006
Human hepatitis B virus enhancer II B1 binding factor (hB1F, also known as LRH-1, hFTF, CPF) belongs to the fushi tarazu factor 1 (FTZ-F1) nuclear receptor subfamily, which was formally designated as NR5A2[1-3]. Like other FsTZ-F1 receptors, hB1F contains a particular FTZ-F1 box which is located at the C-terminus of the DNA-binding domain (DBD) and binds to the response element as monomer[1]. The biological function of hB1F is just being unveiled. It has been reported that hB1F and/or its rodent homologs play an important role in regulating the liver-specific expression of several genes[4,5]. Recent findings pinpoint hB1F as a critical transcription regulator in bile acid biosynthesis[2,6,7], cholesterol homeostasis[8-10], sex hormone biosynthesis[11-13], and lipid metabolism[14].
To facilitate the study on the function of hB1F, we have established 7 transgenic mouse lineages carrying hB1F transgene[15]. In this study, we analyzed the expression of the transgene in livers of these transgenic mouse lines with RT-PCR and Western blotting. Tissue expression pattern of the transgene in one of the transgenic mouse lines was also examined with immunochemical methods. The results of pathological analysis demonstrated that the overexpression of hB1F transgene did not cause pathological changes. We then analyzed the gene expression profile in the liver of transgenic mice with microchip and found that the expression of 3 genes was up-regulated while the expression of 25 genes was down-regulated. Some of the differentially expressed genes were verified with RT-PCR. The expression of farnesyl pyrophosphate synthase, a key enzyme in cholesterol biosynthesis, was inhibited in hB1F transgenic mice.
C57 mice were maintained by Shanghai Nanfang Research Center for Model Organisms (SNRCMO). hB1F transgenic mice were produced in SNRCMO, maintained and bred in the Laboratory Animal Center of the Second Military Medical University.
Total RNA was isolated from tissues with the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Semiquantitative RT-PCR reactions were performed with primer pair sets 5’-CCGACAAGTGGTACATGGAA-3’ and 5’-CTGCTGCGGG TAGTTACA CA-3’ for hB1F cDNA, and 5’-AACTTTGGCATTGTGGAAGG-3’ and 5’-TGTGAGGGAG ATGCTCAGTG-3’ for mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA, which resulted in the generation of 300 bp and 600 bp products, respectively. PCR reactions were performed 30 cycles at 94 °C for 1 min, at 57 °C for 1 min, and at 72 °C for 1 min. PCR products were electrophoresed on 15 g/L agarose gels.
For Western blotting, protein samples from tissues were prepared according to the protocol from Santa Cruz Biotechnology, Inc. Each protein sample (50 μg) was electrophoresed on 100 g/L SDS-polyacrylamide gel and transferred to PVDF membrane. Membranes were blocked with 50 g/L non-fat milk in Tween-TBS (TBST) overnight at 4 °C and incubated with the anti-Flag antibody (Sigma) at a dilution of 1:500 in TBST for 2 h at room temperature. Membranes were washed three times with TBST and incubated with a horseradish peroxidase-conjugated anti-mouse IgG at a dilution of 1:2000 at room temperature for 1 h. Immunodetection was carried out with an enhanced chemilu-minescence kit (Amersham Pharmacia Biotech) according to the manufacturer’s instructions.
Tissue samples were fixed in 10% (vol/vol) neutral formalin, embedded in paraffin, and sectioned for staining. Immunohisto-chemistry was performed on deparaffinized sections. Tissue sections were permeabilized with 3 g/L Triton X-100 in PBS for 30 min. After washed with PBS, sections were saturated for 30 min at room temperature with PBS containing 50 mL/L milk and then incubated for 1 h at room temperature with the anti-Flag antibody (1/250 dilution). This incubation was followed by five washes for 5 min in PBS-10 mL/L milk and then incubated with a sheep anti-mouse IgG (1/100 dilution) in PBS-milk for 1 h at room temperature. Sections were then washed five times for 5 min in PBS and coverslipped with 500 mL/L glycerol in PBS and examined under a microscope and photographed. Immunochemistry and pathological analyses were carrried out at the Department of Pathology, Changhai Hospital of the Second Military Medical University.
RNAs were isolated from livers of two male transgenic mice (TGM-4) and a male C57 mouse. Expressions of 8, 315 genes of the mice were analyzed by using BiostarM-80s cDNA arrays (Biostar genechip Inc., Shanghai, China). Control C57 mouse liver cDNA was labeled with fluorescence Cy3 and TGM-4 liver cDNA was labeled with fluorescence Cy5. Cy3 intensity values were adjusted to Cy3* by multiplying a normalization coefficient. The ratios of Cy5/Cy3* were calculated and genes were identified as either up-regulated when the ratio > 2, or down-regulated when the ratio < 0.5.
Primers used in PCR for CBG gene were 5’-TGTCGTCGCTGCA CTTAATC-3’ and 5’-AGCACATTCCCTTCATCCAG-3’, and for FPPS gene 5’-GGCCATGTGGATCT TGGTAG-3’ and 5’-GAGGAGAGGCTCGTAGCAGA-3’, which resulted in generations of 255 bp and 301 bp products, respectively. The cycling parameters were at 94 °C for 5 min, followed by 30 cycles at 94 °C for 1 min, at 57 °C for 1 min, and at 72 °C for 1 min. The PCR products were separated on 1.5% agarose gels. Signals were quantified by density analysis of the digital images using Alpha image software (Alpha Co., Ltd).
Differences in CBG and FPPS mRNA expressions (comparing CBG/GAPDH or FPPS/GAPDH ratios) were analyzed using one-way ANOVA and by the Student-Newman- Keuls multiple range test.
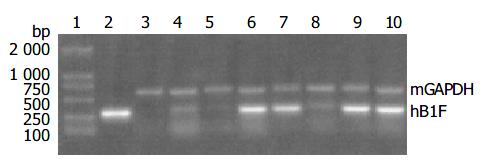
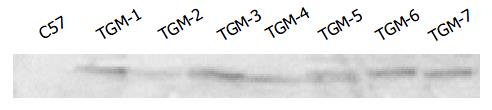
Since the liver is the main organ that expresses hB1F which is involved in the regulation of the liver-specific expression of several important genes, the overexpression of hB1F transgene in mouse liver may serve as an in vivo model to study the function of hB1F in liver. We examined the expression of hB1F transgene in livers of seven transgenic mouse lines. RT-PCR results showed that except TGM-2, six out of seven transgenic mouse lines examined had expression of the transgene (Figure 1). Western blot results demonstated the expression of the transgene was found in livers of all lines with a relatively higher expression in lines TGM-3, TGM-6, and TGM-7 (Figure 2).
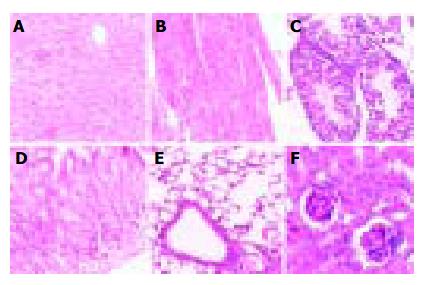
Pathological analysis was performed to examine whether the expression of hB1F transgene might cause any pathological changes in tissues of the transgenic mice. As shown in Figure 3, no obvious pathological change was observed in all tissues from the transgenic mouse line TGM-4. Therefore, as far as the tissues were examined, the expression of hB1F transgene did not result in pathological consequences in TGM-4. Similar results were obtained with other lines (data not shown).
Microchip analysis was perfomed to investigate whether the gene expression profile of the host mice might be altered in the transgenic lines due to the expression of hB1F transgene. The gene expression profiles of cells from the livers of two independent TGM-4 mice were compared with that of the non-transgenic control mouse. Figure 4 represents a visual demonstration of the comparison of gene expression profiles between TGM-4 and C57. The expressions of 28 genes in the livers of TGM-4 mice were found to have changed compared with the C57 control mouse. Among them, 25 genes were down-regulated and 3 genes were up-regulated (Table 1).
| Gene | Ratio 1 | Ratio 2 | Average Ratio | GenBank ID |
| Down-regulated: Trypsin 4 | 0.279 | 0.355 | 0.317 | NM_011646 |
| PAS Ser/Thr kinase | 0.223 | 0.412 | 0.318 | NM_080850 |
| Elongation of very long chain fatty acids-like 3 | 0.274 | 0.402 | 0.338 | NM_007703 |
| 4933406J07Rik RIKEN cDNA | 0.363 | 0.352 | 0.358 | AK016694 |
| Similar to hypothetical protein MGC3169 | 0.367 | 0.372 | 0.369 | BC014728 |
| clone MGC:25675 | ||||
| Farnesyl pyrophosphate synthase | 0.328 | 0.419 | 0.374 | AF309508 |
| Similar to hypothetical protein | 0.422 | 0.328 | 0.375 | BC016095 |
| DKFZp434G2226 clone MGC:27627 | ||||
| T cell immunoglobulin mucin-3 | 0.333 | 0.434 | 0.384 | AF450241 |
| Malate dehydrogenase mitochondrial | 0.419 | 0.350 | 0.385 | NM_008617 |
| 1810009A17Rik RIKEN cDNA | 0.362 | 0.430 | 0.396 | AK007392 |
| Tetratricopeptide repeat domain | 0.386 | 0.412 | 0.399 | NM_009441 |
| 1810007A24Rik RIKEN cDNA | 0.342 | 0.461 | 0.402 | NM_026925 |
| 1810015P03Rik RIKEN cDNA | 0.371 | 0.432 | 0.402 | NM_025458 |
| 4930563E19Rik RIKEN cDNA | 0.424 | 0.394 | 0.406 | AK016201 |
| 1700030E05Rik RIKEN cDNA | 0.400 | 0.422 | 0.411 | BC017608 |
| 2700007P21Rik RIKEN cDNA | 0.364 | 0.458 | 0.411 | AK012215 |
| Capping protein alpha 3 | 0.455 | 0.383 | 0.419 | NM_007605 |
| Trefoil factor 1 | 0.378 | 0.464 | 0.421 | NM_009362 |
| Ketohexokinase | 0.447 | 0.444 | 0.446 | NM_008439 |
| Mus musculus cDNA | 0.429 | 0.467 | 0.448 | AV079172 |
| Amylase 1 salivary | 0.430 | 0.470 | 0.450 | NM_007446 |
| Elastase 2 | 0.495 | 0.448 | 0.472 | NM_007919 |
| Transient receptor protein 2 | 0.446 | 0.499 | 0.472 | NM_011644 |
| Clone IMAGE:3587716 | 0.491 | 0.458 | 0.474 | BC012849 |
| Heavy polypeptide 8 skeletal muscle | 0.486 | 0.489 | 0.488 | M12289 |
| Up-regulated: | ||||
| 602390464F1 Mus musculus cDNA | 2.097 | 2.512 | 2.305 | BG293529 |
| Corticosteroid binding globulin | 2.154 | 2.577 | 2.366 | NM_007618 |
| 602843872F1 Mus musculus cDNA | 3.259 | 2.292 | 2.775 | BG974154 |
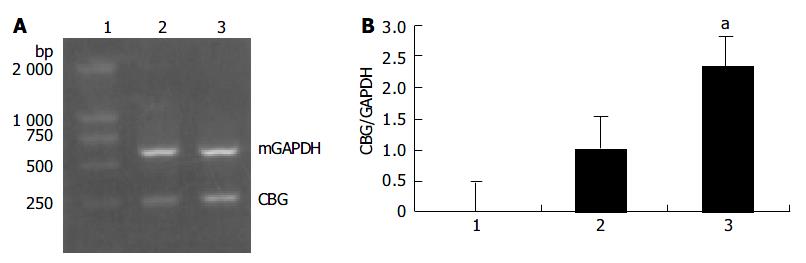
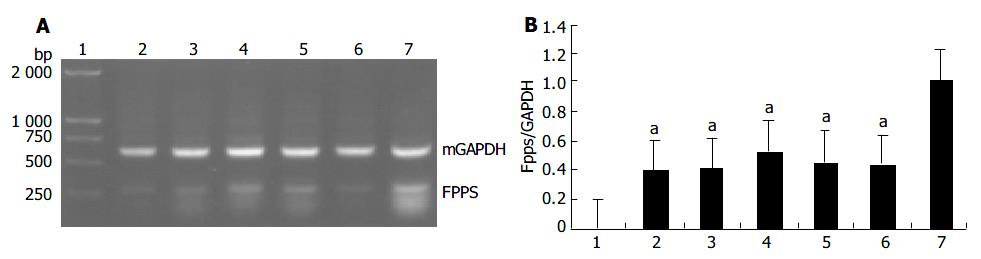
Based on the results of the microchip analysis, some of the differentially expressed genes including corticosteroid-binding globulin (CBG) and farnesyl pyrophosphate synthase (FPPS) were subjected to further analysis. Semi-quantitative RT-PCR was performed with samples used in the microchip analysis and also with additional samples from other transgenic lines, TGM-1, TGM-3, TGM-6, and TGM-7. The corticosteroid binding globulin (serine/cysteine proteinase inhibitor) gene was up-regulated in the liver of TGM-4 (Figure 5) and the farnesyl pyrophosphate synthase gene was down-regulated in all transgenic mouse lines (Figure 6). These results were well consistent with the microchip analysis data.
In this report, we verified the expression of hB1F transgene in several transgenic lines we have constructed previously. Results from RT-PCR and Western blotting analysis indicated that hB1F transgene was expressed in livers of all transgenic lines but with different expression levels. Besides liver, the transgene was also expressed in other organs such as stomach and testis (data not shown). The tissue expression pattern of hB1F transgene was expected since the transcription of thet ransgene was under the control of the CMV early promoter which has a relative broad tissue expression range. Although hB1F was thought previously to be present mainly in liver and pancreas, recent data revealed that it could be expressed in many types of tissues, such as intestine[10,16], ovary[17], adrenal gland[12], and preadipocytes[11,18]. Therefore, the transgenic lines established and verified in this study would provide a valuable animal model for studying the function of hB1F in multiple tissues.
Given that hB1F plays an important role in cholesterol homeostasis and bile acid biosynthesis, it is somewhat unexpected that no discernable pathological changes resulting from the overexpression of hB1F transgene have occurred in these transgenic lines. It is possible that the overexpression of hB1F might negatively feedback on the expression of the endogenous mouse counterpart of hB1F, mLRH-1. It remains to determine whether the expression of the enodgeous mLRH-1 changes in cells overexpressing hB1F. On the other hand, it is apparent from the microchip analysis that the expression of some genes were altered in livers of these transgenic lines. Since disturbance to metabolic pathways such as the cholesterol homeostasis might require a long incubation time before any pathological phenotypes could be observed, it is necessary to perform a long term follow-up investigation on the possible pathological changes.
Among the genes identified to exhibit altered expressions in hB1F transgenic mice, the gene that encodes the farnesyl pyrophosphate synthase (FPPS) is the most interesting one. FPPS could catalyse the formation of farnesyl pyrophosphate (FPP) through the condensation of dimethylallyl pyrophosphate with two molecules of isopentenyl pyrophosphate. FPP is a key cellular intermediate for the biosynthesis of isoprenoids and a precursor of cholesterol, steroid hormones, dolichols, haem A and ubiquinone. Furthermore, it has been found that FPP and its derivative geranylgeranyl pyrophosphate are involved in prenylation, a post-translational modification of a variety of cellular proteins that influence their proper cellular localizations and biological functions[19]. The semi-quantitative RT-PCR results confirmed the down-regulated expression of FPPS in all transgenic lines, suggesting that inhibition of the expression of FPPS has a general effect on hB1F transgenic mice, unrelated to other reasons such as the position effect due to the integration of the transgene. Given the complexity of the regulatory network for the cholesterol homeostasis, it is still early to speculate on the molecular mechanism underlying the inhibition of expression of FPPS in hB1F transgenic mice. Whether hB1F directly or indirectly inhibits the expression of FPPS awaits future study.
Edited by Wang XL Proofread by Xu FM
| 1. | Li M, Xie YH, Kong YY, Wu X, Zhu L, Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem. 1998;273:29022-29031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96:6660-6665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 224] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 3. | Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 831] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 4. | Galarneau L, Paré JF, Allard D, Hamel D, Levesque L, Tugwood JD, Green S, Bélanger L. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853-3865. [PubMed] |
| 5. | Pare JF, Roy S, Galarneau L, Belanger L. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the Hnf3beta, Hnf4alpha, and Hnf1alpha gene promoters. J Biol Chem. 2001;276:13136-13144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1411] [Cited by in RCA: 1515] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 7. | Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1110] [Cited by in RCA: 1137] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 8. | Luo Y, Liang CP, Tall AR. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J Biol Chem. 2001;276:24767-24773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Schoonjans K, Annicotte JS, Huby T, Botrugno OA, Fayard E, Ueda Y, Chapman J, Auwerx J. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 2002;3:1181-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 119] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 10. | Inokuchi A, Hinoshita E, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J Biol Chem. 2001;276:46822-46829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 82] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591-20597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 179] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Wang ZN, Bassett M, Rainey WE. Liver receptor homologue-1 is expressed in the adrenal and can regulate transcription of 11 beta-hydroxylase. J Mol Endocrinol. 2001;27:255-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Sirianni R, Seely JB, Attia G, Stocco DM, Carr BR, Pezzi V, Rainey WE. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J Endocrinol. 2002;174:R13-R17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 14. | Fayard E, Schoonjans K, Annicotte JS, Auwerx J. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J Biol Chem. 2003;278:35725-35731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Wang SL, Yang H, Xie YH, Wang Y, Li JZ, Wang L, Wang ZG, Fu JL. Establishment of transgenic mice carrying the gene of human nuclear receptor NR5A2 (hB1F). World J Gastroenterol. 2003;9:1333-1336. [PubMed] |
| 16. | Rausa FM, Galarneau L, Bélanger L, Costa RH. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3beta in the developing murine liver, intestine and pancreas. Mech Dev. 1999;89:185-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Falender AE, Lanz R, Malenfant D, Belanger L, Richards JS. Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary. Endocrinology. 2003;144:3598-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 132] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Iwaki M, Matsuda M, Maeda N, Funahashi T, Matsuzawa Y, Makishima M, Shimomura I. Induction of adiponectin, a fat-derived antidiabetic and antiatherogenic factor, by nuclear receptors. Diabetes. 2003;52:1655-1663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 560] [Cited by in RCA: 577] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 19. | Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1558] [Cited by in RCA: 1508] [Article Influence: 52.0] [Reference Citation Analysis (0)] |