Published online Oct 15, 2004. doi: 10.3748/wjg.v10.i20.2972
Revised: January 11, 2004
Accepted: February 13, 2004
Published online: October 15, 2004
AIM: To explore the role of hepatitis C virus (HCV) envelope protein 2 (E2) in the induction of apoptosis.
METHODS: A carboxyterminal truncated E2 (E2-661) was transiently expressed in several cultured mammalian cell lines or stably expressed in Chinese hamster ovary (CHO) cell line. Cell proliferation was assessed by 3H thymidine uptake. Apoptosis was examined by Hoechst 33258 staining, flow cytometry and DNA fragmentation analysis.
RESULTS: Reduced proliferation was readily observed in the E2-661 expressing cells. These cells manifested the typical features of apoptosis, including cell shrinkage, chromatin condensation and hypodiploid genomic DNA content. Similar apoptotic cell death was observed in an E2-661 stably expressing cell line.
CONCLUSION: HCV E2 can induce apoptosis in cultured mammalian cells.
- Citation: Zhu LX, Liu J, Xie YH, Kong YY, Ye Y, Wang CL, Li GD, Wang Y. Expression of hepatitis C virus envelope protein 2 induces apoptosis in cultured mammalian cells. World J Gastroenterol 2004; 10(20): 2972-2978
- URL: https://www.wjgnet.com/1007-9327/full/v10/i20/2972.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i20.2972
Hepatitis C virus (HCV) is the major causative agent of post-transfusion and community-acquired hepatitis. HCV has been classified in the Hepacivirus genus within the Flaviviridae family that includes flaviviruses, such as yellow fever and dengue viruses, and animal pestiviruses, such as bovine viral diarrhea virus (BVDV). Infection with HCV usually leads to chronic hepatitis. Although the mechanism for virus persistence is poorly understood, the high mutation rate of viral envelope proteins[1,2] and the suppression of the host immune system[3-5] are believed to contribute to the chronic infection. Analysis of peripheral blood mononuclear cells and liver biopsies from chronic patients suggested that HCV infection could induce apoptosis, which may help the virus escape the immune surveillance and causes liver injuries[6-11]. In vitro studies with either HCV full length RNA[12] or cDNA[13] have demonstrated that apoptosis could be induced by viral proteins. Several independent studies suggested that the viral core protein could induce apoptosis in cultured mammalian cell lines[14,15], while others using similar systems obtained different results[16-18]. Therefore, the viral molecule (s) responsible for the induction of apoptosis has not been clearly identified.
Some members in the Flaviviridae family, e.g. dengue and Langat viruses, could induce apoptosis during infection[19-22]. Duarte dos Santos et al showed that determinants in the envelope protein of dengue type 1 virus could influence the induction of apoptosis[21]. Prikhod’ko et al. demonstrated that apoptosis could be induced by Langat flavivirus infection. Moreover, expression of the viral envelope protein alone was sufficient to induce apoptosis in cultured mammalian cells[22].
Since HCV envelope protein 2 (E2) displays a similar genetic organization as the envelope proteins of these viruses[23], it is possible that E2 may also induce apoptosis. It has been reported that a carboxyterminal truncated E2 (E2-661) without the transmembrane domain is properly folded in cultured mammalian cells[24,25] and has since been used in HCV studies, such as E2-CD81 binding analysis[26] and post-translational processing of E2[27]. In this study, we observed reduced cell proliferations of several cultured mammalian cell lines transiently expressing E2-661. These cells showed the typical features of apoptosis, including cell shrinkage, chromatin condensation and hypodiploid genomic DNA content. Similar apoptotic cell death was observed in an E2-661 stably expressing cell line. This is the first report that HCV E2 can induce apoptosis in cultured mammalian cells.
pSecTagB/sE2 and pSecTagB/sS1E2 containing the insert of E2-661 (aa384-661 of the HCV polyprotein) coding sequence downstream to a signal sequence of Igκ and under the control of the CMV promoter (Figure 1A) were used in the study. To construct pSecTagB/sE2, E2-661 was PCR-amplified from pUC18/E (a gift from Dr. Wang et al[28], Beijing University, GenBank accession# D10934) with primers 5’-GGCGTTAAGCTTAA CACCTACG TG-3’ (HindIII site underlined) and 5’-CAG GAATTCTCACTCTGATCTATC-3’ (EcoRI site underlined). The PCR product was digested with HindIII/EcoRI and cloned in the pSecTagB (Invitrogen). pSecTagB/sS1E2 was constructed by the insertion of a short tag derived from hepatits B virus (HBV) preS1 polypeptide (aa21-47) upstream to the E2-661 coding sequence in pSecTagB/sE2. These constructs were verified by sequencing.
Human liver carcinoma Huh7 and hepatoma HepG2, human cervical carcinoma HeLa and baby hamster kidney BHK-21 cell lines were grown in DMEM supplemented with 100 mL/L fetal calf serum (FCS). Chinese hamster ovary (CHO) cells were grown in DMEM/ F12 (1:1) supplemented with 100 mL/L FCS. The stable E2-661 expressing cell line, CHO/sS1E2 was constructed by transfection of CHO cells with pSecTagB/sS1E2 and selection in DMEM/F12 (1:1) medium containing 400 μg/mL Zeocin (Invitrogen)[24].
Expression plasmids or empty vectors were introduced into cells grown to 60% confluence with LipofectAMINE (Gibco). Transfection efficiency was monitored by co-transfection of pEGFP-C2 (Clontech) which encodes an enhanced green fluorescent protein. Two days after transfection, cells were subjected to Western blot, cell proliferation and apoptosis analysis.
Proteins were resolved on 100 g/L SDS-polyacrylamide gels and electroblotted onto nitrocellulose membranes. Membranes were blocked with 50 mL/L skimmed milk in PBS, followed by incubation with rabbit polyclonal anti-E2 antibody RE2116[29]. The blots were then incubated with horseradish peroxidase conjugated anti-rabbit IgG (Dako) and developed with SuperSignal West Pico stable peroxide solution (Pierce).
CHO and CHO/sS1E2 cells were fixed in ice-cold CMA solution (chloroform: methanol: acetone = 1:2:1), incubated with anti-preS1 monoclonal antibody 125E11[30] 1000-fold diluted in 10 g/L BSA (prepared in PBS) and followed by incubation with FITC conjugated anti-mouse IgG (Santa Cruz). Fluorescence was viewed under a fluorescence microscope.
CHO and CHO/sS1E2 cells were washed twice with PBS and then harvested. Cells were lysed by boiling in the denaturing buffer provided by the manufacturer. Cell lysates were then digested with PNGase F or Endo H (NEB) for 2 h at 37 °C. To analyze the secreted expression products, CHO and CHO/sS1E2 cells were grown in serum free OptiMEM/F12 (1:1) for 12 h. Medium was clarified by centrifugation at 20000 g for 30 min at 4 °C. Expression products were precipitated by an equal volume of ice-cold ethanol, resuspended in a small volume of PBS and subjected to Western blot analysis.
Cell proliferation was measured by 3H thymidine uptake according to a standard protocol[31]. Cells were incubated with 0.05 μCi 3H thymidine (Amersham Pharmacia) for 4 h in complete medium with 100 mL/L FCS. Cells were then washed once with PBS and 100 mL/L trichloroacetic acid (TCA) followed by incubation in 100 mL/L TCA for 10 min at 37 °C. After TCA was removed from the culture dishes, cells were lysed in the lysis buffer containing 0.33 mol/L NaOH and 10 g/L SDS. 3H thymidine incorporation in the cell lysates was measured by liquid scintillation counting.
Apoptosis in stable and transient E2 expressing cells was analyzed by three methods: (1) Hoechst 33258 staining: Cells were seeded on sterile cover glasses placed in the 6-well plates the day before transfection. Two days after transfection, cells were fixed, washed twice with PBS and stained with Hoechst 33258 staining solution according to the manufacturer’s instructions (Beyotime). Stained nuclei were observed under a fluorescence microscope. For the stable cell lines, similar staining procedures were performed without DNA transfection. (2) Flow cytometry: Cells were washed twice with PBS, trypsinized, and resuspended in complete medium with 100 mL/L FCS. Cells were then washed twice again with PBS and fixed with ice-cold 700 mL/L ethanol at 4 °C for 1 h. After the removal of ethanol, cells were incubated in PBS containing 250 μg/mL RNase A and 50 μg/mL propidium iodide (Sigma) at room templeture for 15 min, and stored in the dark at 4 °C until further analysis. Ten thousand cells per sample were analyzed with a FACSCalibur flow cytometer (Beckton-Dickinson). (3) DNA fragmentation: CHO and CHO/sS1E2 cells were washed twice with PBS and harvested. Cells were incubated in lysis buffer [10 mmol/L Tris, 1 mmol/L EDTA, 100 mmol/L NaCl, 5 g/L SDS, 1 μg/μL RNase A, pH8.0] at 37 °C for 30 min. At the end of incubation, proteinase K was added to a final concentration of 0.1mg/mL and the incubation was continued at 55 °C for 4 h. DNA was extracted with phenol/ chloroform and precipitated with ethanol. DNA pellets were dissolved in TE buffer and analyzed on a 20 g/L agarose gel.
Statistical analysis was performed with unpaired 2-sided Student t test. P < 0.05 was regarded as statistically significant.
DNA fragments encoding aa384-661 of the HCV polyprotein (E2-661) were cloned in the expression vector pSecTagB to generate pSecTagB/sE2 and pSecTagB/sS1E2 (Figure1A). Compared to pSecTagB/sE2, E2-661 expressed by pSecTagB/ sS1E2 had a small 27-aa preS1 tag located at the N-terminus to facilitate the characterization of the expression products. The above plasmids were used to transfect Huh7, HepG2, HeLa and BHK-21 cells. Cell lysates were analyzed by Western blot with anti-E2 antibody. E2-661 was detected in E2-661 transient expressing cells (Figure 1B). The expressed protein exhibited a heterogeneous pattern typical of a glycoprotein. The expression of E2-661 could also be detected in the cells (transiently) transfected with pSecTagB/sS1E2 (see below).
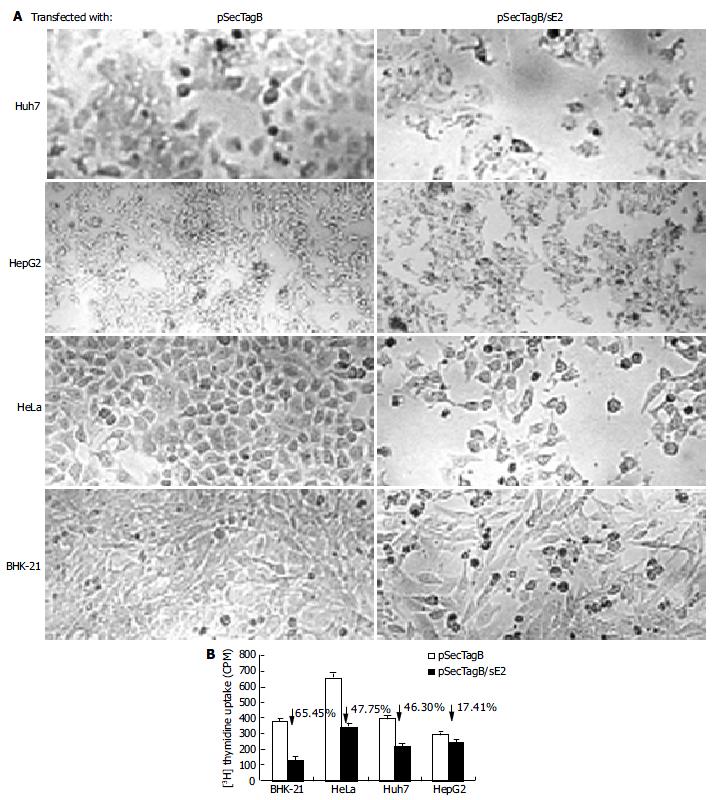
Cells transfected with pSecTagB/sE2 showed significant cell death upon microscope observation (Figure 2A). Reduced cell density, cell rounding and cell shrinkage were the common features of all the cell lines transfected with pSecTagB/sE2. The effect of E2 expression on the cell proliferation was further analyzed by 3H thymidine uptake assay (Figure 2B). Reductions in 3H incorporation were observed with all the cell lines assayed (P < 0.05 for HepG2, P < 0.001 for Huh7, HeLa and BHK-21). Transfection efficiencies were monitored by cotransfection with pEGFP-C2. Since a similar amount of EGFP expressing cells were observed between pSecTagB and pSecTagB/sE2 transfected cells (data not shown), the reduced cell proliferation of E2 expressing cells was not due to the transfection procedure (e.g. transfection reagent and EGFP expression). Similar results were observed with the E2 expressing plasmid pSecTagB/sS1E2 (data not shown). These results suggested that the expression of E2 was responsible for the reduction in cell proliferation.
The reduced proliferation might be due to various reasons. To determine if apoptosis contributed to the reduced cell proliferation of E2 expressing cells, cells transfected with pSecTagB/sE2 or pSecTagB were stained with Hoechst 33258, respectively (Figure 3A). Condensed bright apoptotic nuclei were readily observed amidst the pSecTagB/sE2 transfected cells. The presence of apoptotic cells in E2 expressing cells was further confirmed by flow cytometry. Huh7, HepG2, BHK-21, HeLa cells transfected with pSecTagB/sE2 or control pSecTagB were stained with PI and analyzed by flow cytometry (Figure 3B). Hypodiploid DNA appeared in the sub-G0/G1 region, which represented dead cells. A higher percentage of dead cells was observed in all the four cell lines transfected with pSecTagB/sE2.
E2 induced apoptosis was also investigated in a CHO cell line CHO/sS1E2 stably expressing E2-661. This cell line showed the expression of E2-661 as detected by indirect fluorecence assay with the monoclonal antibody against the preS1 tag (Figure 4A). The fluorescence in CHO/sS1E2 cells suggested the localization of expressed proteins in the cytoplasm. In further characterization by Western blot analysis, cell-associated and secreted E2 proteins (from 105 and 107 cells) were detected with the E2 polyclonal antibody (Figure 4B). The cell-associated E2 protein was sensitive to Endo H (lane 4), suggesting that the glycans carried on it were ER-restricted. On the other hand, secreted E2 was resistant to Endo H (lane 12), suggesting that the glycans on this species of E2 underwent the modification by Golgi enzymes.
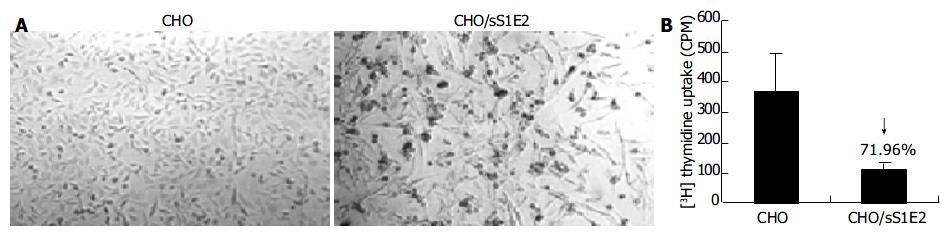
Similar to cells transiently transfected with pSecTagB/sE2, the morphology of CHO/sS1E2 was different from that of the original CHO cell line (Figure 5A). There were obvious differences in cell size and shape between these two cell lines. A significant number of dead cells were readily observed for CHO/sS1E2 and cells could hardly grow to 100% confluency. Similar morphology and growth curve were also observed in other CHO/sS1E2 clones (data not shown), suggesting that they are not the result of clonal selection. 3H incorporation analysis showed that there was a significant reduction (72.0%, P < 0.001) of cell proliferation for CHO/sS1E2 in comparison with CHO cells (Figure 5B).
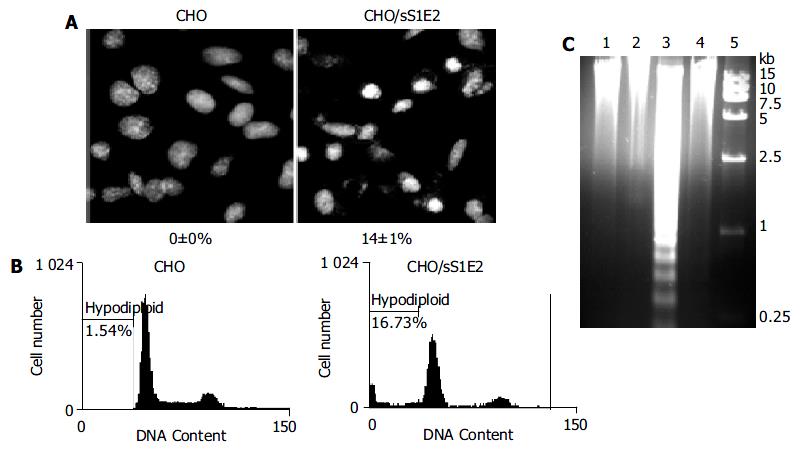
Hoechst 33258 staining of CHO/sS1E2 showed condensed bright nuclei typical of apoptotic dead cells which reached about 14% of the total cells, while almost no apoptotic nuclei were observed in control CHO cells (Figure 6A). Flow cytometry analysis confirmed that there were a significant number of dead cells for CHO/sS1E2 (Figure 6B).
Since the cleavage of chromosomal DNA into fragments of oligonucleosomal size is a biochemical hallmark of apoptosis[32], DNA fragmentation in CHO/sS1E2 was examined by DNA laddering assay. With the freshly seeded CHO/sS1E2 cells, no obvious DNA fragmentation was observed, while clear DNA laddering in -180 bp interval was detected 48 h after cell seeding. No apoptotic DNA fragmentation was observed in CHO cells at either time points (Figure 6C).
In this study, HCV E2 was transiently expressed in several mammalian cells. All tested cells showed shrinkage in morphology and reduced cell proliferation due to the expression of E2. Further evidences, such as condensed chromatin, demonstrated that apoptosis contributed, at least in part, to the cell death induced by E2. E2 induced apoptosis was also observed in a stable E2 expressing cell line CHO/sS1E2. Again there was a significant reduction in cell proliferation of CHO/ sS1E2 in comparison to CHO cells. Hoechst 33258 staining, flow cytometry analysis and DNA laddering assay demonstrated that apoptosis contributed to the reduced cell proliferation. Apoptosis in CHO/sS1E2 was not due to the expression of the small preS1 tag because toxicity was not observed when preS1 was transiently expressed in cultured mammalian cells[33]. In conclusion, our results demonstrate that the expression of HCV E2 could induce apoptosis in cultured mammalian cells.
It has been reported that apoptosis is involved in the pathogenesis of hepatitis C. Immunohistochemical study suggested that the Fas system played an important role in liver injuries of viral hepatitis[34]. There is evidence that immune response (cytotoxic T lymphocyte) might be involved in the apoptosis of hepatocytes in HCV infected patients[35]. Recent studies suggested that apoptosis of hepatocytes might also be due to the cytopathic effect of viral proteins[13,14]. Our result that E2 expression induced apoptosis in cultured mammalian cells including human hepatic cells supports this hypothesis. On the other hand, E2-induced apoptosis may also partly contribute to the escape of HCV from the host immune surveillance. It awaits further investigation whether the expression of E2 can induce apoptosis in lymphocytes.
Studies of Langat flavivirus[22] demonstrated that expression of envelope (E) protein could induce apoptosis via the Caspase 3 pathway. Since HCV E2 and the E protein of Langat flavivirus share similar genetic organization and hydropathy profile[23], it is possible that E2 could induce apoptosis through a similar mechanism. Increased Fas expression observed in HCV patients[7] also suggested the involvement of the caspase-3 pathway. However, comparing the expression levels of secreted and cell-associated E2, we found that in our system, E2 was mainly cell-associated and localized in the cytoplasm. By glycosidase digestion analysis, we found that most of the cell-associated E2 were high-mannose type glycoproteins, which suggested the localization of E2 to the endoplastic reticulum (ER). Many other mammalian expression systems also showed the ER localization of the expressed HCV E2, and a large portion of them formed disulfide-linked aggregates[36-38]. The accumulation of large amount of proteins in the ER might induce ER stress and apoptosis could be triggered via the Caspase 12-mediated apoptosis pathway[39]. Further study is required for understanding the possible pathway of HCV E2 induced apoptosis.
The authors thank Professor Jia-Rui Wu for providing CHO cell line and Professor Yu Wang for the HCV cDNA for this study.
Edited by Wang XL and Xu XQ Proofread by Xu FM
| 1. | Bassett SE, Thomas DL, Brasky KM, Lanford RE. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol. 1999;73:1118-1126. [PubMed] |
| 2. | Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103-126. [PubMed] |
| 3. | Crotta S, Stilla A, Wack A, D'Andrea A, Nuti S, D'Oro U, Mosca M, Filliponi F, Brunetto RM, Bonino F. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J Exp Med. 2002;195:35-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 325] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 4. | Large MK, Kittlesen DJ, Hahn YS. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J Immunol. 1999;162:931-938. [PubMed] |
| 5. | Bain C, Fatmi A, Zoulim F, Zarski JP, Trépo C, Inchauspé G. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology. 2001;120:512-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 307] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 6. | Hiramatsu N, Hayashi N, Katayama K, Mochizuki K, Kawanishi Y, Kasahara A, Fusamoto H, Kamada T. Immunohistochemical detection of Fas antigen in liver tissue of patients with chronic hepatitis C. Hepatology. 1994;19:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 228] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 7. | Calabrese F, Pontisso P, Pettenazzo E, Benvegnù L, Vario A, Chemello L, Alberti A, Valente M. Liver cell apoptosis in chronic hepatitis C correlates with histological but not biochemical activity or serum HCV-RNA levels. Hepatology. 2000;31:1153-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 105] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Emi K, Nakamura K, Yuh K, Sugyo S, Shijo H, Kuroki M, Tamura K. Magnitude of activity in chronic hepatitis C is influenced by apoptosis of T cells responsible for hepatitis C virus. J Gastroenterol Hepatol. 1999;14:1018-1024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Toubi E, Kessel A, Goldstein L, Slobodin G, Sabo E, Shmuel Z, Zuckerman E. Enhanced peripheral T-cell apoptosis in chronic hepatitis C virus infection: association with liver disease severity. J Hepatol. 2001;35:774-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Taya N, Torimoto Y, Shindo M, Hirai K, Hasebe C, Kohgo Y. Fas-mediated apoptosis of peripheral blood mononuclear cells in patients with hepatitis C. Br J Haematol. 2000;110:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Pianko S, Patella S, Ostapowicz G, Desmond P, Sievert W. Fas-mediated hepatocyte apoptosis is increased by hepatitis C virus infection and alcohol consumption, and may be associated with hepatic fibrosis: mechanisms of liver cell injury in chronic hepatitis C virus infection. J Viral Hepat. 2001;8:406-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Kalkeri G, Khalap N, Garry RF, Fermin CD, Dash S. Hepatitis C virus protein expression induces apoptosis in HepG2 cells. Virology. 2001;282:26-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Kalkeri G, Khalap N, Akhter S, Garry RF, Fermin CD, Dash S. Hepatitis C viral proteins affect cell viability and membrane permeability. Exp Mol Pathol. 2001;71:194-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 14. | Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai MM. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691-3697. [PubMed] |
| 15. | Honda M, Kaneko S, Shimazaki T, Matsushita E, Kobayashi K, Ping LH, Zhang HC, Lemon SM. Hepatitis C virus core protein induces apoptosis and impairs cell-cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology. 2000;31:1351-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Marusawa H, Hijikata M, Chiba T, Shimotohno K. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J Virol. 1999;73:4713-4720. [PubMed] |
| 17. | Ray RB, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 188] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 18. | Ray RB, Meyer K, Steele R, Shrivastava A, Aggarwal BB, Ray R. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J Biol Chem. 1998;273:2256-2259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 181] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 19. | Desprès P, Flamand M, Ceccaldi PE, Deubel V. Human isolates of dengue type 1 virus induce apoptosis in mouse neuroblastoma cells. J Virol. 1996;70:4090-4096. [PubMed] |
| 20. | Avirutnan P, Malasit P, Seliger B, Bhakdi S, Husmann M. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J Immunol. 1998;161:6338-6346. [PubMed] |
| 21. | Duarte dos Santos CN, Frenkiel MP, Courageot MP, Rocha CF, Vazeille-Falcoz MC, Wien MW, Rey FA, Deubel V, Desprès P. Determinants in the envelope E protein and viral RNA helicase NS3 that influence the induction of apoptosis in response to infection with dengue type 1 virus. Virology. 2000;274:292-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Prikhod'ko GG, Prikhod'ko EA, Cohen JI, Pletnev AG. Infection with Langat Flavivirus or expression of the envelope protein induces apoptotic cell death. Virology. 2001;286:328-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991;88:5547-5551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 460] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 24. | Wang CL, Zhu LX, Liu J, Zhang ZC, Wang Y, Li GD. Expression and characterization of hepatitis C Virus E2 glycoprotein fused to hepatitis B virus preS1(21-47) fragment in CHO cells. Shengwu Huaxue Yu Shengwu Wuli Xuebao (Shanghai). 2002;34:400-404. [PubMed] |
| 25. | Michalak JP, Wychowski C, Choukhi A, Meunier JC, Ung S, Rice CM, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299-2306. [PubMed] |
| 26. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1572] [Cited by in RCA: 1552] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 27. | Zhu LX, Liu J, Li YC, Kong YY, Staib C, Sutter G, Wang Y, Li GD. Full-length core sequence dependent complex-type glycosylation of hepatitis C virus E2 glycoprotein. World J Gastroenterol. 2002;8:499-504. [PubMed] |
| 28. | Wang Y, Okamoto H, Tsuda F, Nagayama R, Tao QM, Mishiro S. Prevalence, genotypes, and an isolate (HC-C2) of hepatitis C virus in Chinese patients with liver disease. J Med Virol. 1993;40:254-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 68] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Liu J, Zhu L, Zhang X, Lu M, Kong Y, Wang Y, Li G. Expression, purification, immunological characterization and application of Escherichia coli-derived hepatitis C virus E2 proteins. Biotechnol Appl Biochem. 2001;34:109-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Yang HL, Jin Y, Cao HT, Xu X, Li GD, Wang Y, Zhang ZC. Affinity Purification of Hepatitis B Virus Surface Antigen Containing PreS1 Region. Shengwu Huaxue Yu Shengwu Wuli Xuebao (Shanghai). 1996;28:412-417. [PubMed] |
| 31. | Tolleson WH, Melchior WB, Morris SM, McGarrity LJ, Domon OE, Muskhelishvili L, James SJ, Howard PC. Apoptotic and anti-proliferative effects of fumonisin B1 in human keratinocytes, fibroblasts, esophageal epithelial cells and hepatoma cells. Carcinogenesis. 1996;17:239-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 70] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3159] [Cited by in RCA: 3101] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 33. | Hui J, Mancini M, Li G, Wang Y, Tiollais P, Michel ML. Immunization with a plasmid encoding a modified hepatitis B surface antigen carrying the receptor binding site for hepatocytes. Vaccine. 1999;17:1711-1718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Hayashi N, Mita E. Fas system and apoptosis in viral hepatitis. J Gastroenterol Hepatol. 1997;12:S223-S226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Onji M, Kikuchi T, Kumon I, Masumoto T, Nadano S, Kajino K, Horiike N, Ohta Y. Intrahepatic lymphocyte subpopulations and HLA class I antigen expression by hepatocytes in chronic hepatitis C. Hepatogastroenterology. 1992;39:340-343. [PubMed] |
| 36. | Selby MJ, Choo QL, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 37. | Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147-6160. [PubMed] |
| 38. | Dubuisson J, Rice CM. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778-786. [PubMed] |
| 39. | Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935-13940. [PubMed] |