Published online Oct 1, 2004. doi: 10.3748/wjg.v10.i19.2886
Revised: December 15, 2003
Accepted: December 22, 2003
Published online: October 1, 2004
AIM: To study the expression of survivin,a novel member of inhibitors of apoptosis protein (IAP) and its significance in colorectal carcinoma.
METHODS: Survivin mRNA expression was evaluated by semi-quantitative reverse transcriptase polymerase chain reaction (RT-PCR) in 52 colorectal carcinoma samples and 48 adjacent normal colorectal tissue samples. PCR product was sequenced to verify the desired result. Expressions of survivin protein, proliferating cell nuclear antigen labelling index (PI) and apoptotic index (AI) were detected immunohistochemically in 52 human colorectal carcinomas.
RESULTS: The expression of survivin mRNA was detected in a significantly greater proportion of colorectal carcinoma samples than in adjacent normal colorectal tissues (67.3% vs 25%; P < 0.01). There was no relationship between survivin mRNA expression in colorectal carcinomas and sex, tumor size, histological types, lymphnode metastasis, distant metastasis and Dukes’ stage. The PCR product shared 99% of homology with human counterparts. Survivin expression was observed immunohistochemically in 27 of 52 cases of colorectal carcinoma (51.9%). The AI was significently lower in survivin positive group than in survivin negative group (0.67% ± 0.18% vs 1.14% ± 0.42%; P < 0.001), while the PI was greater in survivin positive group than in survivin negative group (51% ± 22% vs 27% ± 18%, P < 0.001).
CONCLUSION: Survivin is a special tumor marker independent of histopathological characteristics. It may play an important role during human colorectal tumorigenesis by inhibiting apoptosis and accelerating proliferative activity of colorectal tumor cells.
- Citation: Chen WC, Liu Q, Fu JX, Kang SY. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol 2004; 10(19): 2886-2889
- URL: https://www.wjgnet.com/1007-9327/full/v10/i19/2886.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i19.2886
Apoptosis is an evolutionally conserved genetic program of cellular suicide and also central to the homeostasis of adult tissues by maintaining the balance between cell production and cell elimination. There is compelling evidence that defects in apoptosis contribute to human diseases such as malignancies. In the evolution of colorectal cancer (CRC) the imbalance between cell proliferation and cell apoptosis may be an important potential factor. A large number of evidences show that many molecules such as p53 and bcl-2 are involved in the regulation of apoptosis during colorectal tumorigenesis. The inhibitor of apoptosis proteins(IAPs), which are widely expressed in all kinds of malignacies, is encoded by the highly-conservative anti-apoptosis gene family and plays an important role in the regulation of apoptosis. IAP molecules contain 1-3 copies of a -70 amino acid zinc-finger fold, and designate the baculovirus IAP repeat (BIR) and their mechanism of action has been directly attributed to the binding of Caspases,thus preventing enzyme activity of the Caspases[1,2]. Among molecules of the IAP family, survivin, a novel anti-apoptosis gene is prominently expressed in CRC and several other malignancies[3,13-18]. The role of survivin in blocking apoptosis is unclear and its ability to inhibit Caspase-3 activity has been widely debated[3-5]. Survivin may paly an important role in the development of CRC. The present study was to examine the expression of survivin in CRC and to evaluate the association between expression of survivin and clinicopathological characteristics of patients with CRC. We also investigated the significence of survivin expression in CRC.
Specimens used for this study were obtained from 52 carcinomas and 48 adjacent normal tissures from the resection margins at the First Affiliated Hospital of Suzhou Univercity. The suspected de novo cancers and patients with a known familial colon cancer syndrome history were excluded. All patients had received neither chemotherapy nor radiation therapy before tumor resection. The diagnosis and histopathologic classification were determined according to the General Rules for Pathologic Studies of National CRC Cooperation Group. In CRC patients, there were 31 males and 21 females, and the mean age of the patients was 61.1 years. According to histological differentiation grade, 52 cases of CRC were classified as well, moderately and poorly differentiated subgroups which included 6, 39 and 7 cases respectively. Seven, 19, 17 and 9 cases were staged as Dukes’ A, B, C and D respectively. After washed with physiological saline, all cancer tissue samples were cut into two pieces by sterile blade.One piece was fixed with formalin for H&E, the other was frozen in liquid nitrogen immediately after the lesion was resected and stored at -80 °C for subsequent assay.
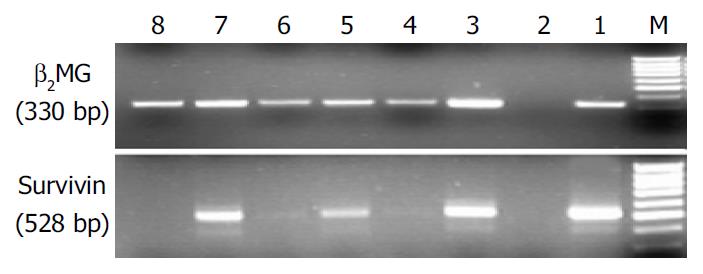
Briefly, total RNAs were extracted from colorectal cancer and normal mucosae, using TRIzol reagent (Life Technologies, Rockville, MD). Prepared RNA (2 µg) was mixed with the random hexamer (100 ng) ,incubated for 5 min at 70 °C, and then quickly chilled at 4 °C for 5 min. RNA samples were reverse-transcriped at 37 °C for 60 min into first-strand cDNA in reverse transcription solution [50 mmol/L Tris-HCl (pH8.3), 40 mmol/L KCl, 8 mmol/L MgCl2, 0.5 mmol/L each dNTP, 225 mg/mL BSA, 5 mmol/L DTT, 25 units of RNasin (Promega, Madison, WI), and 200 units of Mo-MLV reverse transcriptase (Life Sciences, Rockville, MD)] with a total volume of 40 µL. The cDNA samples were incubated at 95 °C for 5 min to inactivate the reverse transcriptase and then chilled. The samples were amplified by adding of 50 µL of PCR mixture [50 mmol/L Tris-HCl (pH8.3), 40 mmol/L KCl, 2 mmol/L MgCl2, 0.2 mmol/L each dNTP, 10 pmoL of each sense and antisense primer, and 1.5 units of Taq polymerase(MBI,)]. Amplification of β2-microglobulin (β2-MG) gene was performed 28 cycles, each for 30 s at 94 °C, for 30 s at 58 °C, and 60 s at 72 °C, followed by an extension at 72 °C for 7 min. Amplification of survivin gene was performed 32 cycles,each for 45 s at 94 °C, 45 s at 61 °C, and for 60 s at 72 °C, followed by an extension at 72 °C for 7 min. The products were electrophoresed on 15 g/L agarose gels and then scanned. Specific primers for survivin gene, targeting a 528-bp fragment, were: 5’-TTGAATCGCGGGACCCGTTGG-3’ (sense) and 5’-CAGAGGCCTCAATCCATGGCA-3’(antisense). As internal standards, β2-microglobulin (β2-MG) gene was used. Primer pairs specific to β2-MG gene, targeting a 330-bp fragment, were used sense: 5’-CTCGCGCTACTCTCTCTTTC-3’ and antisense: 5’-CATGTCTCGATCCCACTTAA-3’.
The PCR products of survivin were isolated and purified using UNIQ-10 purification kit (MBI) and then ligated with pGEM easy T vector by T4 DNA ligase and transformed E. coli JM109. Recombinant vectors were identified through EcoRI digestion and agarose gel electrophoresis. Positive plasmids were sequenced by an ABI PRISM 377 DNA sequencer (Perkin-Elmer Company).
Immunohistochemical staining of survivin was performed by SP method. For PCNA, SABC method was performed. TUNEL assay was used to detect apoptosis. In brief, paraffin sections (4 µm thick) were dewaxed, dehydrated, and then immersed in a 10-3mol/L sodium citrate buffer (pH6.0). For detection of survivin, antigen retrieval was performed by boiling the sections for 20 min in a pressure cooker. For detection of PNCA, the sections were heated in a microwave oven for 10 min to retrieve the antigens. Endogenous peroxidase was inactivated by incubating the sections with 30 mL/L hydrogen peroxide.Nonspecific reactions were blocked by bovine serum. The sections were incubated with a primary antibody overnight at 4 °C. The antibody used was an 8E2 monoclonal antibody(Chemcon, USA) at 1:5 dilution. Rinsed with PBS, the sections were incubated for 30 min at 37 °C with biotinylated secondary antibody and streptavidin conjugated to horseradish peroxidase, respectively. After three rinse with PBS, the sections were incubated with diaminobenzidine substrate, then rinsed with distilled water and counterstained with hematoxylin. As a negative control, PBS was used instead of the primary antibody. PCNA (Maxim, USA) and TUNEL(Roche, USA) assay were performed according to the manufacturer’s instructions.
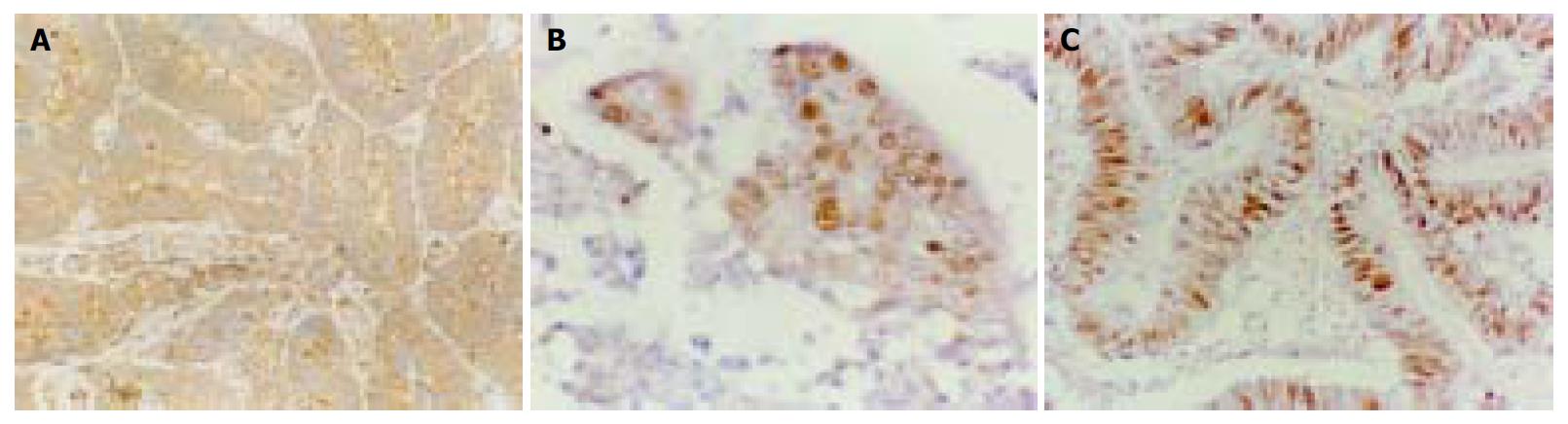
The mean percentage of positive tumor cells for the expression of survivin was determined in at least five areas at 400-fold magnification, and cases with less than 10% positively stained cells were defined as negative. Cases with 10%-29% positively stained cells were defined as “+”, 30%-59% as “++”, and 60% or more as “+++”.These scorings were performed in a blind fashion.
PI and AI were expressed as the ratio of positively stained tumor cells to all tumor cells. If possible, five areas were randomly selected for counting under 400-fold magnification. Otherwise, the whole section underwent assessment.
All results were analyzed by software of SPSS 10.00. P < 0.05 was considered statistically significant.
Fifty two CRC samples and forty eight normal mucosa samples were available for analyses. The expressions of survivvin mRNA were detected in a significantly greater proportion of CRC than in normal mucosa samples (67.3% vs 25% P < 0.01). Survivin mRNA was not detected in normal tissue when the associated cancer was survivin negative (Figure 1).
No relationship was found between survivin mRNA expression in CRC and sex, tumor size, histological types, lymphnode metastasis, distant metastasis and Dukes’ stage (Table 1).
| Variable | Total No | Survivin | P Valur |
| β2MG | |||
| Sex | |||
| Male | 31 | 0.74 ± 0.63 | > 0.05 |
| Female | 21 | 0.77 ± 0.56 | |
| Tumor size | |||
| ≥ 5 cm | 20 | 0.71 ± 0.59 | > 0.05 |
| < 5 cm | 32 | 0.78 ± 0.61 | |
| Histological type | |||
| High | 7 | 0.62 ± 0.51 | > 0.05 |
| Middle | 39 | 0.75 ± 0.59 | |
| Low | 6 | 0.92 ± 0.74 | |
| Lymphnode metastasis | |||
| Positive | 22 | 0.80 ± 0.65 | > 0.05 |
| Negative | 30 | 0.72 ± 0.57 | |
| Distant metastasis | |||
| Positive | 9 | 0.93 ± 0.18 | > 0.05 |
| Negative | 43 | 0.72 ± 0.65 | |
| Duke stage | |||
| A | 7 | 0.55 ± 0.57 | > 0.05 |
| B | 19 | 0.73 ± 0.62 | |
| C | 17 | 0.77 ± 0.73 | |
| D | 9 | 0.93 ± 0.18 |
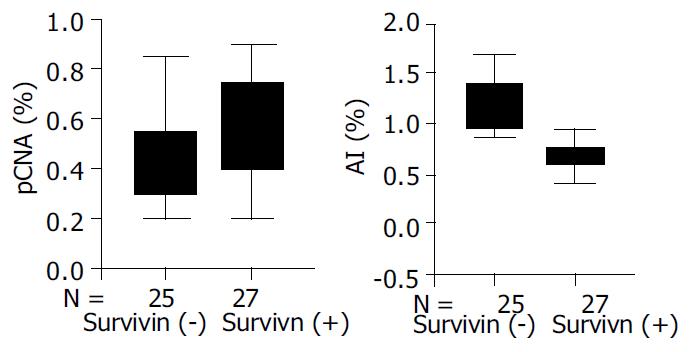
The expression of survivin protein was observed in cytoplasm of malignant tumor cells. Survivin expression was observed immunohistochemically in 27 of 52 cases of colorectal carcinoma (51.9%). To examine whether the expression of survivin correlated with AI and PI, tissure samples obtained from carcinoma were divided into two groups according to the expression of survivin and compared to AI and PI (Figure 2). Survivin positive tumors had significantly lower values for AI than survivin negative tumors (P < 0.001), and PI in survivin positive tumors was higher than in survivin negative tumors (P≤ 0.001, Figure 3).
Hybridization screening of a human P1genomic library with the cDNA of effector cell protease recepor-1 (EPR-1) yielded a new gene spanning -15 kb containing four exons and three introns, and located to band 17q25 by Ambrosini et al[6] in 1997. Survivin is the smallest member of the IAP gene family,structurally characterized by a single BIR module and a -COOH-terminus α-helix coiled domain, substituting a canonical ring finger[6]. Prominently expressed in a variety of apoptosis-regulated organs during embryonic and fatal development[7] , survivin is undetectable in most normal adult tissues,and becomes abundantly reexpressed in a variety of human cancers. The PCR based approach used in our study is exqusitely sensitive and may detect gene transcripts even in a single cell or cell cluster with a comparable specificity. The present study showed that expression of survivin mRNA was detected in a significantly greater proportion of CRC samples than in adjacent normal colorectal tissues (67.3% vs 25%, P < 0.01). The data were consistent with a study by Adida et al[8]. Furthermore, the prevalence of survivin mRNA expression in the present study correlated well with the expression of survivin protein (unpublished data). Recent studies demonstrated that the novel splice variants survivin-△Ex3 and survivin-2B were expressed in malignancies[9,10]. Survivin-△Ex3, which lacks exon 3, exhibited a pronounced anti-apoptotic activity, whereas survivin-2B containing a part of intron 2 as an additional cryptic exon, largely lost its anti-apoptotic activity. These alternative splice variants have not been explored so far. In gastric carcinomas, Mahotka et al[10] showed that survivin-2B might act as a naturally occurring antagonist of survivin and might play a role in tumor progression. We found that CRC also expressed survivin-△Ex3, survivin-2B and survivin. The latter was a dominant transcrip (Figure 1), and shared 99% of homology with human counterparts by sequencing.
The expression of survivin mRNA was independent of clinical pathological characteristics. The result was consistent with previous studies[8,11]. However, in endometrial carcinoma, survivin expression correlated with clinical stage, histological grade, invasive behavior and survival rate[12]. The expression of survivin has been proved as a prognostic mark in non-small cell lung cancer[13], breast carcinoma[14], oesophageal carcinoma[15] and hepatocellular carcinoma[16]. In CRC, Adida et al[8] reported that the predictive value of survivin expression was limited to patients with stage II CRC, those with survivin negative tumors had a five year survival rate of 94.4% compared with 44.8% for patients with survivin positive tumors. Recent studies showed the phenomena of chemo-radioresistance were prevalent in survivin positive tumors and molecular manipulation of survivin expression might enhance chemotherapy and radiation therapy[17-22]. These findings indicate that survivin may play an important role in chemoresistance of cancer cells. However, the clinical importance of survivin expression remains unclear in patients with cancer.
We found that survivin positive tumors had significantly lower values for AI than survivin negative tumors, and PI was higher in survivin positive tumors than in survivin negative tumors (P < 0.001 for both). These findings indicate that the overexpression of survivin not only inhibits cell death but also reflects the presence of a greater number of proliferating cells. By inhibiting apoptosis and promoting proliferation, survivin plays an important role in colorectal tumorigenesis.
There has been a considerable interest in survivin biomedical researches. The exploitation of survivin might provide important predictive and prognostic clues in diagnosis, and offer new therapeutic alternatives for cancer. Further studies are required to identify the potential role and mechanism of survivin in the development of CRC.
Edited by Wang XL and Xu FM
| 1. | Miller LK. An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol. 1999;9:323-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 267] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 2. | Altieri DC. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol Med. 2001;7:542-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 324] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 3. | Zhu XD, Lin GJ, Qian LP, Chen ZQ. Expression of survivin in human gastric carcinoma and gastric carcinoma model of rats. World J Gastroenterol. 2003;9:1435-1438. [PubMed] |
| 4. | Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001;276:3238-3246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 336] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 5. | Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 527] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 6. | Shankar SL, Mani S, O'Guin KN, Kandimalla ER, Agrawal S, Shafit-Zagardo B. Survivin inhibition induces human neural tumor cell death through caspase-independent and -dependent pathways. J Neurochem. 2001;79:426-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2342] [Cited by in RCA: 2383] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 8. | Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43-49. [PubMed] |
| 9. | Sarela AI, Macadam RC, Farmery SM, Markham AF, Guillou PJ. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut. 2000;46:645-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 248] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 10. | Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097-6102. [PubMed] |
| 11. | Krieg A, Mahotka C, Krieg T, Grabsch H, Müller W, Takeno S, Suschek CV, Heydthausen M, Gabbert HE, Gerharz CD. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002;86:737-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Lin LJ, Zheng CQ, Jin Y, Ma Y, Jiang WG, Ma T. Expression of survivin protein in human colorectal carcinogenesis. World J Gastroenterol. 2003;9:974-977. [PubMed] |
| 13. | Takai N, Miyazaki T, Nishida M, Nasu K, Miyakawa I. Survivin expression correlates with clinical stage, histological grade, invasive behavior and survival rate in endometrial carcinoma. Cancer Lett. 2002;184:105-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 78] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Monzó M, Rosell R, Felip E, Astudillo J, Sánchez JJ, Maestre J, Martín C, Font A, Barnadas A, Abad A. A novel anti-apoptosis gene: Re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol. 1999;17:2100-2104. [PubMed] |
| 15. | Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127-134. [PubMed] |
| 16. | Ikeguchi M, Kaibara N. survivin messenger RNA expression is a good prognostic biomarker for oesophageal carcinoma. Br J Cancer. 2002;87:883-887. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Ikeguchi M, Hirooka Y, Kaibara N. Quantitative analysis of apoptosis-related gene expression in hepatocellular carcinoma. Cancer. 2002;95:1938-1945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 18. | Asanuma K, Moriai R, Yajima T, Yagihashi A, Yamada M, Kobayashi D, Watanabe N. Survivin as a radioresistance factor in pancreatic cancer. Jpn J Cancer Res. 2000;91:1204-1209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 152] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 19. | Ikeguchi M, Liu J, Kaibara N. Expression of survivin mRNA and protein in gastric cancer cell line (MKN-45) during cisplatin treatment. Apoptosis. 2002;7:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Kato J, Kuwabara Y, Mitani M, Shinoda N, Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J. Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer. 2001;95:92-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 21. | Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177-11182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 315] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 22. | O'Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103-13107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 497] [Article Influence: 19.9] [Reference Citation Analysis (0)] |