INTRODUCTION
Hepatitis B virus (HBV) belongs to the family hepadnaviridae which replicate their genome by reverse transcription, and causes acute and chronic hepatitis and is strongly associated with the development of primary hepatocellular carcinoma (HCC)[1,2]. HBV has a unique open reading frame, with the sequence being highly conserved among different mammalian hepadnaviruses, coding for a 16.5 kDa protein known as hepatitis B virus X protein (HBx). X gene is a multifunctional regulatory factor which has been identified as a potential oncogene. Transcriptional activation of diverse cellular genes by the HBx has been suggested as one of the mechanisms for HBV-associated HCC[2]. HBx has been shown to activate a wide variety of cellular and viral genes in trans activation including the HBV enhancers, tumor suppressor p53[3,4], and proto-oncogenes c-jun, c-fos and c-myc[5,6]. HBx-responsive elements also include nuclear factor kappa B (NF-κB), activator protein-1 (AP-1), activator protein-2 (AP-2) and cAMP-response element (CRE)[7,8]. Since HBx has no ability to bind to dsDNA, protein-protein interaction seems to be crucial for HBx transactivation. The interaction of HBx with cellular proteins may trigger a cascade of phosphorylation and dephosohorylation events which might lead to a general up-regulation of gene expression[9]. Identification of cellular X-interactive proteins would provide insights into the mechanism of HBV celluar effects. Several technigues have been used to study possible protein-protein interaction with HBx. Using these approaches, HBx has been discovered to interact with many proteins[9]. Nonetheless, to the author’s knowledge, the functional significance of these interactions has not yet been elucidated. In the present study, we screened a clone encoding a novel X-interactive protein which was homologous to Homo sapiens cytochrome C oxidase III (cox III) from normal human liver cDNA library by the Saccharomyces cerevisiae two-hybrid system.
MATERIALS AND METHODS
Plasmids and cells
Plasmids pAS2-1, PCL1, PLAM5’-1, normal human liver pACT2-cDNA library and Saccharomyces cerevisiae AH109 were purchased from Clontech, USA. Saccharomyces cerevisiae AH109 was grown in YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose). This yeast strain carried LacZ, HIS3 and ADE2 reporter genes under the control of Gal4-binding site and was used to screen the liver cDNA library. Escherichia coli JM105 was provided as a gift from Research Institute of Hepatology, Beijing University, China.
Reagents
Restriction enzymes (EcoR I and Pst I), TaqDNA polymerase and T4 DNA ligase were purchased from Promega, USA. DNA gel extraction kit was provided by Jingmei Company, China. Gal4 DNA-BD monoclonal antibody, matchmaker AD LD-insert screening amplimers and yeast culture medium were purchased from Clontech, USA. Glass beads (acid-washed) were purchased from Sigma, USA. Alkaline phosphatase-conjugated goat anti-mouse IgG was purchased from Wuhan Boster Biological Technology Company, China.
pAS2-1-X construction
The X region of the HBV genome was amplified by PCR with XF and XR as forward and reverse primers which contained an EcoR I site and a Pst I site, respectively, for convenience of cloning. The sequences of the primers with the restriction enzyme sites underlined were: XF, 5’-ACGGAATTCATGGCTGCTAGGCTGTG-3’; XR, 5’-ATCCTGCAGAGGTGAAAAAGTTGCAT-3’. The fragment amplified was from nucleotides 1 374 to 1838 on HBV genome. The template for this reaction was extracted from sera of HBV DNA-positive patients. The reaction mixture was subjected to 30 cycles of PCR amplification. Each cycle included denaturation at 94 °C for 30 s, annealing at 58 °C for 1 min, and extension at 72 °C for 30 s , and a final extension for 7 min at 72 °C. The approximate 464 bp fragment was digested by EcoR I and Pst I, and recombined into the plasmid pAS2-1 by T4 DNA ligase. The reconstructed plasmid was subsequently named pAS2-1-X and identified by PCR and auto-sequencing assay. Auto-sequencing assay was performed in Bioasia Biologic Technology Company and the resulting sequence was analyzed in the database of EMBL\GeneBank by the BLAST program.
Transformation of pAS2-1-X into AH109
Yeast AH109 was transformed with pAS2-1-X by lithium acetate-mediated method demonstrated by Gietz et al[10] and plated in selective SC/-trp medium. Plasmids were isolated from transformants and X gene was amplified. Filter assay was used to exclude the auto-activation function of pAS2-1-X in AH109. Expressed X-BD fusion protein in yeasts transformed with PAS2-1-X was detected by Western blotting. The cells were collected by centrifugation and yeast protein extract was prepared according to urea/SDS method. SDS-polyacrylamide gel electrophoresis was performed and protein extracts were electroblotted onto nitrocellulose membrane. After being blocked with nonfat milk, the membrane was added to 1:3000 diluted Gal4 DNA-BD monoclonal antibodies and incubated for 1 h, after washed with TBST. Then, the secondary alkaline phosphatase-conjugated goat anti-mouse IgG was added and incubated for 2 h. The proteins were visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium. Protein extracts of untransformed yeast cells were used as negative control.
Screening of human liver cDNA library by the yeast two-hybrid system
The screening procedure used here was a modification of the method described by Gietz et al[10]. Yeast cells were transformed with pAS2-1-X and pACT2-cDNA library by lithium acetate-mediated method , plated in selective SC/-trp-leu-his-ade medium, incubated at 30 °C for 7 d and selected for histidine, leucine and tryptophan prototrophy. The freshly growing clones were assayed for β-gal activity by replicaplating the yeast transformants onto Whatman filter paper. The filters were then snap frozen twice in liquid nitrogen for 10 s and incubated in a buffer containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside solution at 30 °C for 1-8 h. Positive interactions were detected by appearance of blue clones. Plasmids PCL1 and PLAM5’-1 were transformated into AH109 as positive and negative controls, respectively. Segregation analysis was performed to eliminate false positive clones and thus true positive clones were obtained.
Analysis of positive clones
pACT2-cDNA plasmid genome was isolated following the method described by Gietz et al[10]. Briefly, the true positive clones were incubated with SC/-leu liquid medium at 30 °C overnight, followed by spinning down the cells by centrifuging at 14000 g for 5 min, and resuspension of pellets in lysis buffer (20 g/L Triton 100, 10 g/L SDS, 10 mmol/L NaCl, 10 mmol/ L Tris-HCl, pH8.0, 1 mmol/L Na2EDTA) was added with phenol, chloroform and isoamyl alcohol (volume fraction 25:24:1). Then the suspension was vortexed vigorously with acid-washed glass beads, lysate was centrifugated and plasmid DNA was harvested. The pACT2-cDNA plasmid was purified by CsCl gradient centrifugation to permit PCR using the matchmaker AD LD-insert screening amplimers which annealed to GAL4-AD. The PCR reaction consisted 30 amplification cycles, each cycle included denaturation at 94 °C for 30 s, annealing at 68 °C for 3 min, and a final extension for 7 min at 72 °C. Auto-sequencing assay was performed in Shanghai Sangon Biological Engineering Technology and Service Corporation, the resulting sequences were compared against the databases of EMBL\GenBank by the BLAST program.
Mating experiment
pAS2-1-X, pAS2-1, pLAM5’-1 transformed yeasts in SC/-trp medium and positive clones were incubated in SC/-leu medium at 30 °C for 2 d, respectively, then removed. Two types of transformants were incubated into YPD medium at 30 °C for 4 h. Finally, transformants were subcultured in SC/-leu-trp medium, incubated at 30 °C for 2 d and assayed for β-gal activity.
RESULTS
Identification of the pAS2-1-X
The fragment of approximately 460 bp was amplified from the reconstructed plasmid pAS2-1-X, and auto-sequencing assay confirmed the fragment inserted into plasmid pAS2-1 had a high identity (99.9%) with X gene.
Detection of the transformed yeast AH109
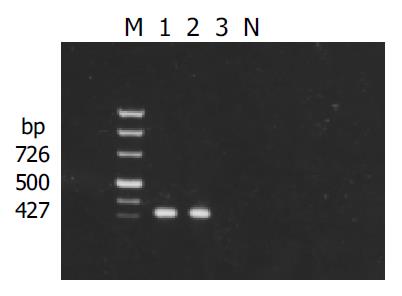
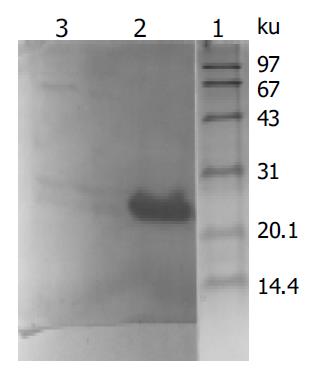
X gene was amplified from the plasmid isolated from the transformed yeast cells (Figure 1). Filter assay excluded the auto-activation function of pAS2-1-X in the cells. Staining for X-BD was obtained in yeast cells transformed with pAS2-1-X (Figure 2). pAS2-1-X could be used as a bait plasmid in yeast two-hybrid system.
Figure 1 Result of detection of X gene in yeasts transformed with pAS2-1-X.
M: PCR marker; lanes 1 and 2: transformed yeasts; lanes 3: untransformed yeasts; N: negative control.
Figure 2 Western blot analysis of expressed fusion protein in yeasts transformed with PAS2-1-X.
Lane 1: protein moclecular weight standard; lane 2: protein extracted from transformed yeasts; lane 3: protein extracted from untransformed yeasts.
Screening of the liver cell cDNA library
Of the 5 × 106 transformants screened, 103 were grown in the selective SC/-trp-leu-his-ade medium, 19 were positive for β-gal activity, and only 1 clone passed through the segregation analysis.
Analysis of positive clones
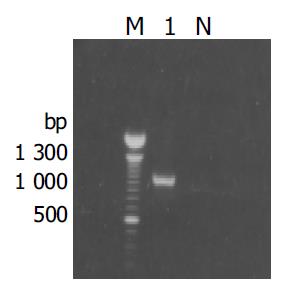
The fragment of approximately 900 bp was amplified from the true positive clones (Figure 3). DNA sequence analysis of the fragment revealed that the yeast plasmid insert had a high identity (98%) with the Homo sapiens cox III gene (GeneBank ID:BC013930).
Figure 3 Results of amplification for cDNA fragment by PCR from the positive clones.
M: 100 bp DNA ladder; lane1: positive clone; N: negative control.
Mating experiment
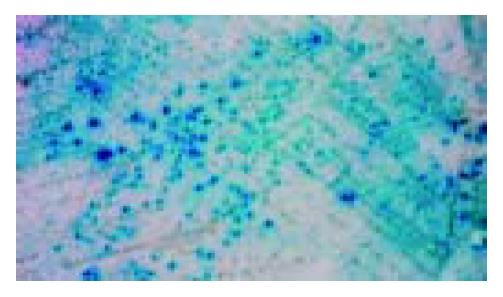
The transformants containing pAS2-1-X and positive pACT2-cDNA gave a blue colour by filter assay, whereas the transformants containing pAS2-1 or pLAM5’-1 and positive pACT2-cDNA gave a white colour. Mating experiment confirmed the specific interaction between positive clones and HBx (Figure 4).
Figure 4 Mating experiment for the interaction between cox III and X protein in yeast cells.
DISSCUSION
Chronic HBV infection is strongly associated with the development of HCC, but the mechanisms by which HBV induces events leading to the genesis of HCC remains unclear. Several studies have suggested a possible role of HBx in the process of HCC, a few studies have reported transformation of certain cell types in vitro by HBx[11] and initiation of HCC in HBx transgenic mice[12,13], but its direct oncogenicity seems to depend on the strength, and duration of protein expression and the genetic background of mice and cells. It is more likely that HBx causes transactivation of cell regulatory genes or interacts with one or more cellular proteins to initiate the development of human HCC through upregulation of cell growth and mutagenesis. The transactivation function of HBx has been shown to involve both direct interaction with transcriptional factors, such as RPB5 of RNA polymerases[14], TATA-binding protein[15] or activating transcription factor (ATF) /CRE-binding protein (CREB)[16] and activation of signal transduction pathways, such as ras/raf/MAP-kinase[17], protein kinase C (PKC)[18] and SAPK/JNK[19,20]. HBx is predominatly localized in the cytoplasm with a low level of nuclear distribution, whereas recent findings from two independent laboratories indicated that HBx could localize at the mitochondria. Confocal laser microscopy of hepatoma cells transfected with respective expression vectors showed colocalization of the third member of the family of human genes that encode the voltage-dependent anion channel (HVDAC3) and HBx in mitochondria. Moreover, stable cationic fluorophore dye showed that HBx expression in hepatoma cells led to alteration of mitochondrial transmembrane potential, an abnormal aggregation of mitochondrial structures, release of cytochrome C from mitochondria, decrease of mitochondrial membrane potential and membrane blebbing of cells, which are characteristics of cell death. These functional roles of HBx in affecting mitochondrial physiology are associated with HBV-induced liver injury and development of HCC[21,22].
Since HBx seems to accomplish its functions via protein-protein interaction, identification of the HBx- interactive cellular proteins represents a major goal in order to define the function of HBx in HBV replication and liver carcinogenesis. The yeast two-hybrid system using a genetic approach, offers a way to clone and identify genes that interact with a protein of interest through in vivo complementation in yeast cells[23,24]. In the present study, we screened and cloned a novel HBx-interactive protein from a normal human liver cDNA library. Sequence analysis revealed it was the homolog of Homo sapiens cox III. The interaction of HBx with cox III was confirmed by yeast-two hybridization system 3. The growth of yeast cells harboring both pAS-1-X and pACT2-cox III recombinant plasmids in His-independent medium, formation of blue-colonies detected by the β-gal assay, and the behaviors of the cells in false-positive elimination tests suggested that the binding of cox III was HBx specific. Mating experiment further identified the interaction between cox III and HBx in yeast cells.
Cytochrome C oxidase (COX), the terminal enzyme of the mitochondrial respiratory chain, catalyzes the transfer of electrons from reduced cytochrome C to molecular oxygen. cox III, which is encoded by the mitochondrial DNA and synthesized within the mitochondria, represents one of the large subunits of COX and acts as the catalytic core of the enzyme[25]. Defective COX activity resulting in deficient adenosine triphosphate generation may contribute to the development of Alzheimer’s disease[26], cardiomyopathy[27], neonatal giant cell hepatitis[28] and other clinically heterogeneous diseases[29,30]. Incorrect assembly of the critical subunits incluing cox I,cox IIand cox III, is a major mechanism leading to COX deficiencies. COX also plays a key role in inducing apoptosis in multiple cell types through transcriptional activation of the respective genes[31]. Mitochodrial dysfuction and transcriptional function changes have been identified to be associated with chronic liver disease and oncogenic processes[32]. Kuo et al[33] reported the integration of HBV with human hepatoma cells might cause mitochondrial defects in the ATP synthase 6 and cox III genes. Studies on the effects of HBx on apoptotic pathways indicated that HBx blocked the actirit, of caspases 8, 9 and 3 and the release of cytochrome c, it also upregulated PKC, extracellular signal-regulated kinase (ERK), phosphatidyl inositol-3-kinase (PI-3-K) and NF-κB activity[34]. The study of NF-κB and HBx expression in human HCC tissues detected by immunohistochemistry SP method strongly suggested that NF-κB was abnormally activated in HCC, which was probably related to HBx. The ability of HBx to induce the activation of NF-κB has been demonstrated by mobility shift and reporter gene expression assays with lysates from HBx-transfected HepG2 cells[35]. Furthermore, HBx was shown to bind to a voltage-dependent anion channel and alter the mitochondrial transmembrane potential and consequently induce activation of transcription factors, which include STAT-3 and NF-κB[21]. Cogswell et al[36] reported that the NF-κB regulatory pathway existed in mitochondria and NF-κB could negatively regulate the expression of both cox III and cytochrome B mRNAs. Whether and how the interaction of HBx with cox III regulates the activity of transcription factor NF-κB has remained obscure. However, based on our data, we hypothesize that HBx might impair mitochondrial respiration chain and energy metabolism by combining cox III.
Future investigations will focus on the identification of cox III properties and physiological significance of interaction of HBx with cox III and probably with other mitochondrial proteins. Further inquiries in this area may pave the way for investigation of possible mechanisms leading to chronic infection and subsequent progression to HCC in the context of mitochondrial association of HBx.