Published online Sep 15, 2004. doi: 10.3748/wjg.v10.i18.2735
Revised: December 24, 2003
Accepted: January 15, 2004
Published online: September 15, 2004
AIM: To study the expression of a disintegrin and metalloproteinase 17 (ADAM17) mRNA in hepatocellular carcinoma (HCC) and to evaluate the relationship between ADAM17 mRNA expression and clinicopathological features of HCC.
METHODS: Hepatocellular carcinomas (HCC) from 31 cases were divided into small HCC (SHCC), nodular HCC (NHCC) and solitary large HCC (SLHCC) according to tumor diameter and the number of nodes. ADAM17 mRNA expressions were compared among those groups by means of semi-quantitative reverse transcription polymerase chain reaction (RT-PCR). The relationship between ADAM17 mRNA expression level and clinicopathological features of HCC was evaluated.
RESULTS: NHCC had lower differentiation and was more frequently of microvascular invasion (10/12) than SHCC (3/11) and SLHCC (3/8) (P < 0.05), but no statistical difference was observed between SHCC and SLHCC comparing their clinicopathological features. ADAM17 mRNA expression was detected in 77.4% (24/31) of HCC tissues and was significantly higher than that in paired non-cancerous liver tissues in which only 35.5% (11/31) of the samples were detected of the expression (P < 0.05). The expression of ADAM17 mRNA was much higher in NHCC than in SHCC and SLHCC (P < 0.05), while no significant difference was discovered between SHCC and SLHCC. The quantities of ADAM17 mRNA were significantly higher in poorly differentiated HCC than in well or moderately differentiated HCC, but no statistical difference was found concerning liver cirrhosis, tumor capsule formation or microvascular invasion of the cancer.
CONCLUSION: The increased expression of ADAM17 may play a key role in the development of HCC. The expression levels of ADAM17 mRNA varied among different pathological types of HCC. Lower mRNA expression of ADAM17 mRNA in SLHCC may be associated with the better molecular pathological features of SLHCC.
- Citation: Ding X, Yang LY, Huang GW, Wang W, Lu WQ. ADAM17 mRNA expression and pathological features of hepatocellular carcinoma. World J Gastroenterol 2004; 10(18): 2735-2739
- URL: https://www.wjgnet.com/1007-9327/full/v10/i18/2735.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i18.2735
Hepatocellular carcinoma (HCC) is one of the most common malignant diseases in the world. Approximately 560000 new cases of HCC are diagnosed each year, constituting 6% of all new human cancers[1]. The HCC mortality rate in China is approximately 20.4/100000 and is the second leading cause of cancer death among Chinese males[2]. The prognosis of large hepatocellular carcinoma (LHCC) is generally considered to be worse than small hepatocellular carcinoma (SHCC), but long-term survival of some LHCC, especially solitary large hepatocellular carcinoma (SLHCC), could be frequently observed after curative resection[3-5]. From the clinical observation and our research, we hypothesized that SLHCC was of relatively better biological behavior[2]. Furthermore, we have preliminarily proved our hypothesis in a series of researches by showing that SLHCC was of relatively better pathological features and surgical prognosis[2,3,6]. Although many systemic studies have been performed, the unique clinical and molecular pathological features of SLHCC are still far from a clear deep understanding and need further investigation.
A disintegrin and metalloproteinase 17 (ADAM17) is a kind of transmembrane metalloproteinase[7], which can cleave the ectodomain of many transmembrane proteins. The substrates of ADAM17 mediated cleavage include tumor necrosis factor-α (TNF-α), tumor necrosis factor receptor type I, (TNFR I) and tumor necrosis factor receptor type II (TNFR II), interleukin1 receptor type II (IL1R II), Notch receptor, L-selectin, mucin1 (MUC1), CD30, tumor necrosis factor-related activation-induced cytokine (TRANCE) and many ligands of epidermal growth factor receptor (EGFR), such as transforming growth factor-α (TGF-α), heparin-binding epidermal growth factor-like growth factor (HB-EGF), amphiregulin and so on[8-19]. Hassan reported that the oxygen radicals generated by smoke stimulated ADAM17 to cleave transmembrane amphiregulin. The binding of amphiregulin to EGFR then promoted proliferation of lung cancer cells[20]. The cleavage function of ADAM17 was required for the activation of EGFR by TGF-α in Hela cells, and the high expression level of ADAM17 in mammary tumors was correlated with high activation rate of EGFR[21]. Another experiment showed that single metastatic lung cancer cell in bone marrow presented overexpression of ADAM17[22]. All these suggested that ADAM17 played a key role in the development of some cancers. It is well known that the EGFR mediated pathway played an important role in HCC development[23,24]. The overexpression of EGFR and its ligands were observed in HCC tissue and were related to the prognosis of HCC patients[25,26]. However, much less is known about the mechanisms involved in signal transduction, which activate the EGFR in HCC. It is even not sure whether there is an increased activation rate of EGFR in HCC besides the overexpression of EGFR and its ligands. As ADAM17 played an important role in the activation of EGFR[27] and up to now, there is no report about the expression and function of ADAM17 in HCC, so to have a better understanding of the molecular mechanisms involved in HCC development and the unique molecular pathological features of SLHCC, we studied the expression of ADAM17 in HCC and paired non-cancerous liver tissues.
Thirty-one fresh HCC specimens (26 from males and 5 from females) were obtained by surgical resection in Xiangya Hospital, Central South University between May 2002 and February 2003. Tissues 1 cm away from the edge of the cancer were used as the paired non-cancerous control. The tissues used for RNA extraction were immediately snap-frozen in liquid nitrogen and stored at -70 °C. The specimens used for pathological study were fixed with 10% formalin, dehydrated by conventional methods, embedded in paraffin, cut into slices of 5 μm thick and stained by haematoxylin and eosin.
The specimens were divided into SHCC group (largest diameter less than 5 cm for single tumor nodule or the sum of diameters less than 5 cm for two tumor nodules, n = 11), SLHCC group (single tumor nodule with largest diameter more than 5 cm, n = 8) and NHCC group (two or more tumor nodules in the liver, only two tumor nodules and the sum of diameters less than 5 cm were excluded, n = 12).
All specimens were examined under a microscope after haematoxylin-eosin staining by two pathologists. Four aspects of clinicopathological features including liver cirrhosis, Edmondson classification, capsule formation and microvascular invasion were studied.
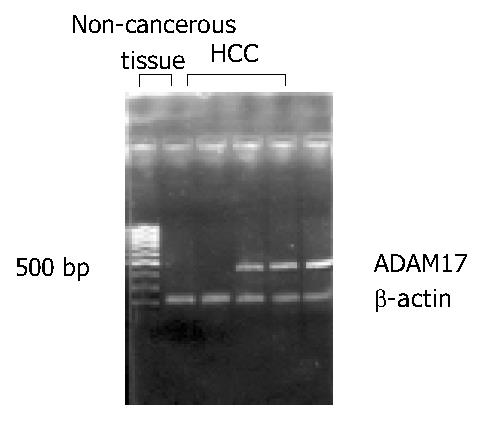
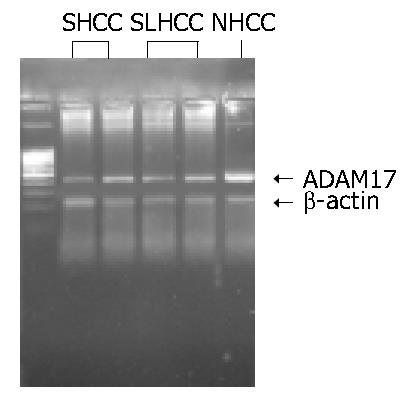
The total RNA was extracted from HCC tissue and paired non-cancerous liver tissue by using the TRIzol (Invitrogen, USA). The quality of RNA was checked through the ribosomal RNA bands on the gel. Two µg of each intact total RNA sample was reverse-transcribed to complementary DNA (cDNA) by using RT-PCR kit (MBI, USA). The PCR primer sequences used were as follows: sense: 5’-GCACAGGTAATAGCAGTGAGTGC-3’ and antisense: 5’-CACACAATGGACAAGAATGCTG-3’ for ADAM17; sense: 5’-TTCCAGCCTTCCTTCCTGG-3’ and anti-sense: 5’-TTGCGCTCAGGAGGAGCAAT-3’ for β-actin. The sizes of PCR products were 440 bp for ADAM17 and 218 bp for β-actin. The procedure was as follows: denaturation at 94 °C for 5 min, then 40 cycles of denaturation at 94 °C for 50 s; annealing at 52 °C for 1 min and extension at 72 °C for 1 min. The PCR products were electrophoresed in 20 g/L agarose gels, and visualized under ultraviolet light. The expression ratio of ADAM17 mRNA to β-actin was determined by Eagle Eye II photo-analysis system.
The χ2 test was used for quantitative enumeration data. The student’s t test and one-way analysis of variance were used for qualitative data. The statistic analysis was performed by statistical software SPSS11.0. P value less than 0.05 was considered significant.
According to the results of pathological study (shown in Table 1), the NHCC group had higher incidences of microvascular invasion (10/12) compared with SLHCC (3/8) and SHCC (3/11) (P < 0.05). Only 8.3% NHCC (1/12) was classified as Edmondson I-II, while 62.5% SLHCC (5/8) and 63.6% SHCC (7/11) were classified as Edmondson I-II. The differentiation of NHCC was significantly poorer than SLHCC and SHCC (P < 0.05). The other two pathological features of SLHCC and SHCC were also better than NHCC but the difference did not reach statistical significance. No statistical difference of the four pathological features was observed between SLHCC and SHCC.
As shown in Figure 1, RT-PCR products of ADAM17 and β-actin presented at the same site as previously designed. The expression of ADAM17 mRNA was detected in 77.4% (24/31) of HCC tissues, much higher than that in paired non-cancerous liver tissues in which only 35.5% (11/31) tissues were detected of the expression (P < 0.05).
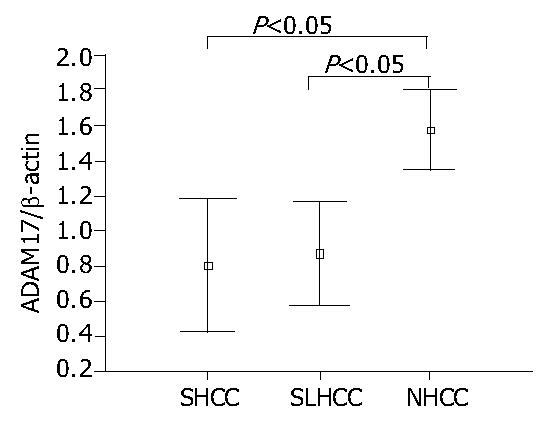
As shown in Figure 2, Figure 3, the transcription level of ADAM17 was 0.8 ± 0.7 (mean ± SD) in SLHCC and 0.9 ± 0.6 in SHCC. Both were significantly lower than that in NHCC (1.6 ± 0.5) (P < 0.05). No statistical difference in transcription level of ADAM17 was observed between SLHCC and SHCC.
The transcription level of ADAM17 was compared between different pathological types of HCC. The ADAM17 transcription level was much higher in HCC samples classified as Edmondson I-II compared with those classified as Edmondson III-IV (P < 0.05). While no statistical difference in transcription level of ADAM17 was observed when capsule formation, liver cirrhosis, and microvascular invasion were concerned (Table 2).
| Sample (n) | Transcription level of ADAM17 (mean ± SD) | P | |
| Liver cirrhosis | |||
| Present | 22 | 1.141 ± 0.743 | > 0.05 |
| Absent | 9 | 0.996 ± 0.662 | |
| Microvascular invasion | |||
| Present | 16 | 1.279 ± 0.648 | > 0.05 |
| Absent | 15 | 0.893 ± 0.750 | |
| Capsule formation | |||
| Present | 18 | 1.058 ± 0.157 | > 0.05 |
| Absent | 13 | 1.187 ± 0.800 | |
| Edmondson classification | |||
| I-II | 13 | 0.768 ± 0.204 | < 0.05 |
| III-IV | 18 | 1.349 ± 0.157 |
HCC ranks fifth in frequency worldwide among all malignancies[1]. Surgery is the only potential curative treatment of HCC[28]. The post-operative survival of SHCC was generally considered to be better than LHCC, but long-term disease-free survival of some LHCC was also frequently observed after curative resection. Furthermore, some clinical investigation showed that the 5-year survival rate of LHCC after curative resection was not statistically different from SHCC[4,5]. This kind of LHCC was named SLHCC because it was of some unique characters such as: isolated lesion, expanding growth and relatively integrated fibrous capsule formation et al[2]. Previous studies showed that SLHCC had relative better prognosis after curative resection[2,3,6]. Consistent with this, the relatively better pathological features of SHCC were found in this study. The differentiation of SLHCC was much better than that of NHCC, and NHCC was more frequently of microvascular invasion compared with SLHCC. No statistic difference in pathological features was observed between SHCC and SLHCC.
ADAM17 is a sheddase of many transmembrane proteins. The substrates of ADAM17 mediated shedding include many ligands of EGFR such as TGF-α, HB-EGF, amphiregulin et al[14]. It has been well known that the activation of EGFR is essential for the carcinogenesis and metastasis of many cancers, while ADAM17 mediated shedding is a key mechanism of sending signals to activate EGFR[19-21]. Overexpression of ADAM17 has been observed in gastric tumor, mammary cancer, leukemia cell lines and prostate cancer cell lines[20,21,29-32]. In this study the transcription of ADAM17 was detected in 77.4% (24/31) HCC samples and was statistically higher than that in paired non-cancerous samples in which only 35.5% (11/31) were detected (P < 0.05). This suggests that ADAM17 may play a key role in HCC development.
Previous studies showed that SLHCC had relative better prognosis after curative resection. To understand the underlying molecular mechanism, the transcription levels of ADAM17 in SLHCC, SHCC and NHCC were also studied. As expected, the transcription level of ADAM17 was much higher in NHCC than in SLHCC and SHCC, while no statistical difference was observed between SLHCC and SHCC. The transcription level of ADAM17 mRNA was also detected to be statistically higher in samples classified as Edmondson III-IV compared with those classified as I-II. The higher expression of ADAM17 mRNA in NHCC as well as in poorly differentiated HCC suggested that it might facilitate tumor invasiveness. The lower transcription level of ADAM17 in SLHCC was probably associated with the relatively better molecular pathological features of SLHCC.
The relationship between ADAM17 mRNA expression and the other two tumor features: microvascular invasion and tumor capsule formation was also studied in this study. Although much higher ADAM17 mRNA level was detected in tumors with microvascular invasion and those without integrated capsule formation, the difference did not reach statistical significance. This maybe due to the relatively small sample size in this study. As only 31 samples were used in the study, especially after the samples were divided into three groups, the significance of the research results may be impaired. The role of ADAM17 in HCC invasiveness needs further investigation with a larger sample size.
The increased transcription of ADAM17 may facilitate the growth and invasiveness of HCC in several ways. (1) Overexpressions of EGFR and its ligands were observed in HCC and were related to the prognosis. However, ADAM17 mediated shedding is a key mechanism of sending signals to activate EGFR. ADAM17 is required for the activation of EGFR by TGF-alpha or amphireglin in several cancer cells and the high expression of ADAM17 in mammary tumors was correlated with a high activation rate of EGFR[20,21]. As ADAM17 regulates the ligands production and activity of EGFR, the overexpression of ADAM17 is probably as important as the increased expression of EGFR and its ligands for HCC development; (2) ADAM17 can function as an effector of G protein-coupled receptor (GPCR)-mediated signaling. Activation of GPCR specifically results in ADAM17 cleavage and release of amphiregulin, which could activate EGFR and regulate the proliferation and motility of squamous cell carcinoma[32]. Overexpression of GPCR was also observed in HCC and was strongly correlated with carcinogenesis of HCC[33]. ADAM17 may be a key element of communication between GPCR and EGFR in HCC and facilitates HCC development; (3) TRANCE could activate osteoclast and help cancer cells metastasize to bones. Overexpression of TRANCE was found in bone metastatic lesion of several cancers[34]. ADAM17 may play a role in the bone metastasis of HCC by cleaving TRANCE; (4) The shedding of TNFR by ADAM17 may cause disorder of host immune system as the soluble form of TNFR could bind to the TNF and block its attack to cancer cells[35].
Other transmembrane proteins associated with HCC metastasis such as Fas ligand, CXCL12, E-cadherin, interleukin-6α[36-39] were also the suspected substrates of ADAM17 mediated shedding, so it is of particular significance to study the sheddase role of ADAM17 in HCC development. The ADAM metalloproteinase family includes more than 30 members by now[40]. Many of them not only function as metalloproteinases to shed transmembrane proteins[41-43] in cancer but also can work as adhesion molecules[44,45]. In contrast to the matrix metalloproteinases, relatively few data on expression of ADAMs in cancer tissue are available. The expression of ADAM12 and ADAM9 were recently studied in HCC tissue and were of particular importance[46]. To study the role of other ADAM family members may help us better understand HCC development.
Edited by Chen WW Proofread by Zhu LH and Xu FM
| 1. | Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1727] [Cited by in RCA: 1710] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 2. | Yang LY, Huang GW. Surgical strategy of large hepatocellular carcinoma. Linchuang Waike Zazhi. 2001;9:4-5. |
| 3. | Yang LY, Huang GW, Huang JH, Yang JQ, Lü XS, Han M. Surgical excision of 114 cases of large hepatocellular carinomas. Zhongguo Shiyong Waike Zazhi. 2002;22:353-355. |
| 4. | Poon RT, Fan ST, Wong J. Selection criteria for hepatic resection in patients with large hepatocellular carcinoma larger than 10 cm in diameter. J Am Coll Surg. 2002;194:592-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 126] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 5. | Régimbeau JM, Farges O, Shen BY, Sauvanet A, Belghiti J. Is surgery for large hepatocellular carcinoma justified. J Hepatol. 1999;31:1062-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Yang LY, Huang GW, Huang JH, Yang JQ. Resection of large hepatocellular carcinoma without emplyment of pringle maneuver. Zhonghua Gandan Waike Zazhi. 2003;9:331-333. |
| 7. | Black RA. Tumor necrosis factor-alpha converting enzyme. Int J Biochem Cell Biol. 2002;34:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 261] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 8. | Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2422] [Cited by in RCA: 2420] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 9. | Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275:14608-14614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 400] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 10. | Condon TP, Flournoy S, Sawyer GJ, Baker BF, Kishimoto TK, Bennett CF. ADAM17 but not ADAM10 mediates tumor necrosis factor-alpha and L-selectin shedding from leukocyte membranes. Antisense Nucleic Acid Drug Dev. 2001;11:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Slack BE, Ma LK, Seah CC. Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-alpha converting enzyme. Biochem J. 2001;357:787-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Wang X, He K, Gerhart M, Huang Y, Jiang J, Paxton RJ, Yang S, Lu C, Menon RK, Black RA. Metalloprotease-mediated GH receptor proteolysis and GHBP shedding. Determination of extracellular domain stem region cleavage site. J Biol Chem. 2002;277:50510-50519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Thathiah A, Blobel CP, Carson DD. Tumor necrosis factor-alpha converting enzyme/ADAM 17 mediates MUC1 shedding. J Biol Chem. 2003;278:3386-3394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 156] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 14. | Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J Biol Chem. 2002;277:12838-12845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 339] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 15. | Garton KJ, Gough PJ, Blobel CP, Murphy G, Greaves DR, Dempsey PJ, Raines EW. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1). J Biol Chem. 2001;276:37993-38001. [PubMed] |
| 16. | Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 833] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 17. | Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000;275:10379-10387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 246] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 18. | Lum L, Wong BR, Josien R, Becherer JD, Erdjument-Bromage H, Schlöndorff J, Tempst P, Choi Y, Blobel CP. Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J Biol Chem. 1999;274:13613-13618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 301] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 19. | Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1255] [Cited by in RCA: 1259] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 20. | Lemjabbar H, Li D, Gallup M, Sidhu S, Drori E, Basbaum C. Tobacco smoke-induced lung cell proliferation mediated by tumor necrosis factor alpha-converting enzyme and amphiregulin. J Biol Chem. 2003;278:26202-26207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Borrell-Pagès M, Rojo F, Albanell J, Baselga J, Arribas J. TACE is required for the activation of the EGFR by TGF-alpha in tumors. EMBO J. 2003;22:1114-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 226] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 22. | Klein CA, Seidl S, Petat-Dutter K, Offner S, Geigl JB, Schmidt-Kittler O, Wendler N, Passlick B, Huber RM, Schlimok G. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotechnol. 2002;20:387-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 220] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 23. | Hisaka T, Yano H, Haramaki M, Utsunomiya I, Kojiro M. Expressions of epidermal growth factor family and its receptor in hepatocellular carcinoma cell lines: relationship to cell proliferation. Int J Oncol. 1999;14:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Miyaki M, Sato C, Sakai K, Konishi M, Tanaka K, Muraoka M, Kikuchi-Yanoshita R, Nadaoka Y, Kanda H, Kitagawa T. Malignant transformation and EGFR activation of immortalized mouse liver epithelial cells caused by HBV enhancer-X from a human hepatocellular carcinoma. Int J Cancer. 2000;85:518-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 25. | Daveau M, Scotte M, François A, Coulouarn C, Ros G, Tallet Y, Hiron M, Hellot MF, Salier JP. Hepatocyte growth factor, transforming growth factor alpha, and their receptors as combined markers of prognosis in hepatocellular carcinoma. Mol Carcinog. 2003;36:130-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 105] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Ito Y, Takeda T, Sakon M, Tsujimoto M, Higashiyama S, Noda K, Miyoshi E, Monden M, Matsuura N. Expression and clinical significance of erb-B receptor family in hepatocellular carcinoma. Br J Cancer. 2001;84:1377-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 237] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 27. | Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE, Castner BJ, Gerhart MJ, Paxton RJ, Black RA. TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase. Ann N Y Acad Sci. 2003;995:22-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 145] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 28. | Yu AS, Keeffe EB. Management of hepatocellular carcinoma. Rev Gastroenterol Disord. 2003;3:8-24. [PubMed] |
| 29. | Yoshimura T, Tomita T, Dixon MF, Axon AT, Robinson PA, Crabtree JE. ADAMs (a disintegrin and metalloproteinase) messenger RNA expression in Helicobacter pylori-infected, normal, and neoplastic gastric mucosa. J Infect Dis. 2002;185:332-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | McCulloch DR, Harvey M, Herington AC. The expression of the ADAMs proteases in prostate cancer cell lines and their regulation by dihydrotestosterone. Mol Cell Endocrinol. 2000;167:11-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997;235:437-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 88] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 32. | Gschwind A, Hart S, Fischer OM, Ullrich A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003;22:2411-2421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 269] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 33. | Yamamoto Y, Sakamoto M, Fujii G, Tsuiji H, Kenetaka K, Asaka M, Hirohashi S. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 164] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Huang L, Cheng YY, Chow LT, Zheng MH, Kumta SM. Tumour cells produce receptor activator of NF-kappaB ligand (RANKL) in skeletal metastases. J Clin Pathol. 2002;55:877-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Wallach D, Engelmann H, Nophar Y, Aderka D, Kemper O, Hornik V, Holtmann H, Brakebusch C. Soluble and cell surface receptors for tumor necrosis factor. Agents Actions Suppl. 1991;35:51-57. [PubMed] |
| 36. | Ethell DW, Kinloch R, Green DR. Metalloproteinase shedding of Fas ligand regulates beta-amyloid neurotoxicity. Curr Biol. 2002;12:1595-1600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Marin V, Montero-Julian F, Grès S, Bongrand P, Farnarier C, Kaplanski G. Chemotactic agents induce IL-6Ralpha shedding from polymorphonuclear cells: involvement of a metalloproteinase of the TNF-alpha-converting enzyme (TACE) type. Eur J Immunol. 2002;32:2965-2970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 38. | Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276:4972-4980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 217] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 39. | Scotton CJ, Wilson JL, Scott K, Stamp G, Wilbanks GD, Fricker S, Bridger G, Balkwill FR. Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Cancer Res. 2002;62:5930-5938. [PubMed] |
| 40. | Brachvogel B, Reichenberg D, Beyer S, Jehn B, von der Mark K, Bielke W. Molecular cloning and expression analysis of a novel member of the Disintegrin and Metalloprotease-Domain (ADAM) family. Gene. 2002;288:203-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 41. | Moss ML, Lambert MH. Shedding of membrane proteins by ADAM family proteases. Essays Biochem. 2002;38:141-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Gutwein P, Oleszewski M, Mechtersheimer S, Agmon-Levin N, Krauss K, Altevogt P. Role of Src kinases in the ADAM-mediated release of L1 adhesion molecule from human tumor cells. J Biol Chem. 2000;275:15490-15497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 148] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 43. | Blobel CP. Functional and biochemical characterization of ADAMs and their predicted role in protein ectodomain shedding. Inflamm Res. 2002;51:83-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Thodeti CK, Albrechtsen R, Grauslund M, Asmar M, Larsson C, Takada Y, Mercurio AM, Couchman JR, Wewer UM. ADAM12/syndecan-4 signaling promotes beta 1 integrin-dependent cell spreading through protein kinase Calpha and RhoA. J Biol Chem. 2003;278:9576-9584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 89] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 45. | Zhu Pc, Sun Y, Xu R, Sang Y, Zhao J, Liu G, Cai L, Li C, Zhao S. The interaction between ADAM 22 and 14-3-3zeta: regulation of cell adhesion and spreading. Biochem Biophys Res Commun. 2003;301:991-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 46. | Le Pabic H, Bonnier D, Wewer UM, Coutand A, Musso O, Baffet G, Clément B, Théret N. ADAM12 in human liver cancers: TGF-beta-regulated expression in stellate cells is associated with matrix remodeling. Hepatology. 2003;37:1056-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 161] [Article Influence: 7.3] [Reference Citation Analysis (0)] |