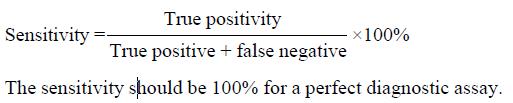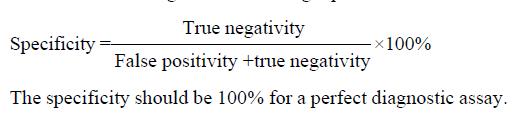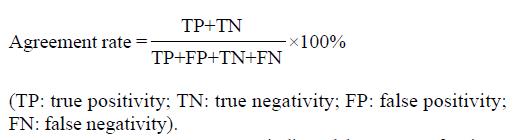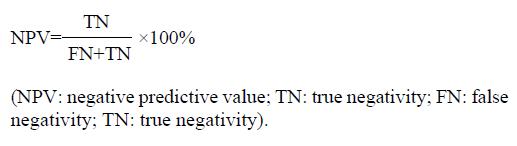Published online Aug 15, 2004. doi: 10.3748/wjg.v10.i16.2439
Revised: September 18, 2003
Accepted: October 7, 2003
Published online: August 15, 2004
AIM: To design and establish a method of multiplex PCR normalization for simultaneously detecting HBV and HCV.
METHODS: Two pairs of primers with a 20 bp joint sequence were used to amplify the target genes of HBV and HCV by two rounds of amplification. After the two rounds of amplification all the products had the joint sequence. Then the joint sequence was used as primers to finish the last amplification. Finally multiplex PCR was normalized to a single PCR system to eliminate multiplex factor interference. Four kinds of nucleic acid extraction methods were compared and screened. A multiplex PCR normalization method was established and optimized by orthogonal design of 6 key factors. Then twenty serum samples were detected to evaluate the validity and authenticity of this method.
RESULTS: The sensitivity, specificity, diagnostic index and efficiency were 83.3%, 70%, 153.3% and 72.2%, respectively for both HBsAg and anti-HCV positive patients, and were 78.6%, 80%, 258.6% and 79.2%, respectively for HBsAg positive patients, and were 75%, 90%, 165% and 83.3%, respectively for anti-HCV positive patients.
CONCLUSION: The multiplex PCR normalization method shows a broad prospect in simultaneous amplification of multiple genes of different sources. It is practical, correct and authentic, and can be used to prevent and control HBV and HCV.
- Citation: Wang N, Gao XQ, Han JX. Simultaneous detection of HBV and HCV by multiplex PCR normalization. World J Gastroenterol 2004; 10(16): 2439-2443
- URL: https://www.wjgnet.com/1007-9327/full/v10/i16/2439.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i16.2439
Multiplex PCR uses several pairs of primers that target different genes to simultaneously amplify several different target sequences at a high speed and with a high efficiency. As many amplification factors may interact and produce non-specific amplification, its clinical use is limited. According to the characteristics of a small fragment that did not complement to the genes of PCR primers, we established a multiplex PCR normalization method (This method has been patented in the Chinese Patent Agency). By creating a primary reaction that was appropriate to all target templates, the multiplex was normalized to a single target PCR. This method could overcome the difficulties in establishing and optimizing the multiplex reaction system. In this article the practicality and effectiveness of multiplex PCR were validated by simultaneous detection of HBV and HCV.
Twenty-eight serum samples were collected from Jinan Central Hospital and Jinan Infectious Disease Hospital. The samples were all validated by ELISA method, in which 14 cases were HBsAg ( + ), 8 were anti-HCV ( + ), and 6 were positive for both HBsAg and anti-HCV. Ten cases were negative for both HBsAg and anti -HCV and used as control.
HBV PCR kits and HCV PCR kits were purchased from Institutes of Liver Diseases, Peking University Medical College. dNTPs, AMV, proteinase K, isothiocyanate guanidine and Triton X –100 were products of TAKARA. PCR Amplification MinicyclerTM was from MJ Research, USA. Innotech ImagerTM 2200 was from Alpha Innotech Incoporation.
Method 1[1]: Proteinase K (10 mg/mL) was added to 150 μL serum and incubated at 50 °C for 2 h. RNA and DNA were extracted with phenol-chloroform at pH4.0 and 8.0 respectively, and then nucleic acids were precipitated with Isopropyl alcohol. The final pellets were dissolved in 10 μL RNase free water and stored at -70 °C.
Method 2[2]: 200 μL guanidine isothiocynate and 20 μL glass powder were added to 200 μL serum and incubated at ambient temperature for 90 min and centrifuged at 12000 g. The supernatant was removed and the pellet was washed and dissolved in reverse transcription buffer PCR detection.
Method 3[3]: 90 μL lytic buffer (120 g GUSCN, 100 mL 0.1mol/L Tris-HCl, pH6.4), 0.2 mol/L EDTA (20 μL, pH8.0) and glass powder were added to 50 μL serum and incubated at room temperature for 10 min, mixed and centrifuged at 12 000 g for 15 s, washed 2 times with washing solution (120 g GUSCN, 100 mL 0.1 mol/L Tris-HCl, pH 6.4, 2.6 g Triton × 100), and further washed with 700 mL/L ethanol and acetone. Then acetone was removed and dried at 56 °C for 10 min and TE buffer was added to the pellets and incubate at 56 for 2 min and then centrifuged for 2 min. The supernatant was transferred to another tube for further amplification.
Method 4[4]: 200 μL lytic buffer[6 mol/L guanidine hydrochloride, 10 mmol/L Tris-HCl (pH 7.5), 200 g/L Triton X-100 (pH 4.4)], and 10 mmol/L urea, were added to 200 μL serum, proteinase K 40 μL, glass powder 10 μg, and incubated in 72 °C water bath for 10 min, then 100 μL Isopropyl alcohol was added and centrifuged at 8000 r/min for 1 min. The supernatant was removed, and washed with 100 μL washing solution [20 mmol/L NaCl, 20 mmol/L Tris-HCl (pH7.5) and 1000 mL/L ethanol] and centrifuged at 8000 r/min for 1 min. The supernatant was removed and washed and centrifuged at 8000 r/min for 1 min and 13000 g for 10 s, dried and dissolved in 50 μL RNase free water.
The primers for the first round of PCR: HBV reverse primer: 5’-GAT GAT GGG ATG GGA ATA CA-3’ (position: 2566-2586 of P gene), HBV RT primer: 5’-GCT GGT TCA CAT TGT GAG GGG AGT CTA GAC TCG TGG TGG A-3’ with the former 20 bp as the joint sequence, and the latter 20 bp in the position of the overlap of genome P region and Pre-S region (position 2921-2941). There was a 395 bp between R and RT primers. HCV reverse primer: 5’-ATC ACT CCC CTG TGA GGA A-3’ which was located between 47-65 bp. RT –PCR primer: 5’-GCT GGT TCA CAT TGT GAG GGC TAC GAG ACC TCC CGG GGC A-3’. The former 20 bp was the joint sequence, the latter 20 bp was in 313-332 of HBV genome. The product between R and RT primers was a 315 bp fragment. The second round primers: N primer sequence was 5’-GCT GGT TCA CAT TGT GAG GG TAG AGG ACA AAC GGG CAA CA3’, the former 20 bp was the joint sequence, and the latter 20 bp was in 2703-2723 of HBV genome. RT-PCR primer was the same as that of the first round, the amplification product was a 278 bp fragment between N and RT. The HCV N primer was 5’-GCT GGT TCA CAT TGT GAG GG GGG AGA GCCAT AGT GGT CTG-3’, the former 20 bp was the joint sequence, and the latter 20 bp was located between 132-151 of HCV. RT primers were the same as the first round of amplification and the product was a 241 bp fragment. After two rounds of amplification, the third round was amplified with the common primers T: 5’GCT GGT TCA CAT TGT GAG GG3’. The primers were synthesized by phosphoamidite method with 391 DNA synthesizer. The synthesized primers were purified with OPC column[5].
Sensitivity indicated the percent of positive cases by the diagnostic assay in the patient group. It was calculated according to the following formula.
Math 1
Specificity indicated the percent of negative cases determined by the diagnostic assay in control group. It was calculated according to the following equation.
Math 2
Diagnostic index was a combined parameter to evaluate the sensitivity and specificity. It was the sum of sensitivity and specificity. The perfect diagnostic index should be 200%, and it should not be lower than 100%.
Agreement rate indicated the ability of a diagnostic assay to correctly discern the patients and control cases. It was calculated by the following method. The perfect agreement rate of a diagnostic assay should be 100%.
Math 3
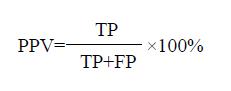
Positive predictive value (PPV) indicated the percent of patients in the positive results of diagnostic assay. It was calculated according to the following equation.
Math 4
Negative predictive value (NPV) indicated the percent of non-patients in the negative results of diagnostic assay. It was calculated according to the following equation.
Math 5
Four methods were used to extract HBV DNA from HBsAg positive sera of patients. The results of amplification were compared to that of ELISA (Table 1).
| Method | Sample number | |||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| ELISA | + | + | + | + | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + |
| Method 1 | - | - | + | + | + | - | + | - | - | - | - | + | - | - | - | - | - | - | - | - |
| Method 2 | + | - | - | + | + | + | + | + | + | - | + | + | + | - | - | - | + | + | - | - |
| Method 3 | + | + | + | + | + | + | + | + | + | - | - | + | + | + | + | - | + | + | + | + |
| Mrthod 4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + |
The positive amplification rates were 20%, 60%, 85% and 95% for methods 1 to 4 respectively.
Fifteen anti-HCV positive sera were extracted with the four methods and the results were compared with ELISA (Table 2).
| Method | Sample number | ||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| ELISA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Method 1 | - | - | - | - | - | - | - | - | + | - | - | - | - | - | - |
| Method 2 | - | - | - | + | - | - | - | - | - | - | - | - | - | - | - |
| Method 3 | - | + | + | + | - | - | - | - | + | + | - | - | + | - | - |
| Mrthod 4 | + | + | + | + | + | + | + | + | + | + | - | + | + | + | + |
The positive amplification rates were 6%, 6%, 30% and 95% for methods 1 to 4, respectively compared to the ELISA.
Six samples positive for both HBsAg and anti-HCV were extracted and amplified, the simultaneous extraction positive rate was 100% for HBV and 83% for HCV, respectively.
Orthogonal method was used to select the six common factors affecting the amplification, which were the concentrations of dNTPs, Mg2+, Taq polymerase, AMV and primers (including HBV and HCV). Five levels were designed for each factor. The pattern of orthogonal was L 25[5,6]. The PCR reaction was as follows: HBV, 8 μL HCV template, 5 μL 10 × buffer, 40 U RNase inhibitor, and other components and finally ddH2O was added to make the total volume 50 μL. Twenty-five kinds of amplification system were available. After paraffin oil was added, reactions were performed at 50 °C for 30 min, at 95 °C for 2 min,and 30 cycles at 95 °C for 30 s, at 55 °C for 30 s, at 72 °C for 1 min. Then 10 μL amplification product was identified by 15 g/L agarose electrophoresis. The results showed under the condition of protocol 13, that both HBV and HCV targets were successfully amplified, therefore protocol 13 was chosen as the first round protocol. The amplification program was at 50 °C for 30 min, at 95 °C for 2 min, and at 95 °C for 30 s, at 55 °C for 30 s and at 72 °C for 1 min for a total of 30 cycles.
According to the orthogonal method and experience, 6 protocols were designed. The reaction condition was at 95 °C for 30 s, at 55 °C for 30 s, and at 72 °C for 45 s for a total of 10 cycles. The results showed that the two target bands of HBV and HCV were successfully separated in protocol 1. Therefore the protocol 1 was chosen as the final optimal protocol.
According to the results of the above two rounds of amplification, 7 protocols were designed for the third round of amplification. All the amplification reactions were done at 95 °C for 30 s, at 55 °C for 30 s, at 72 °C for 1 min for a total of 30 cycles. The results showed that the HBV and HCV target bands were discerned more obviously under the condition of protocol 2, which was therefore determined as the optimal protocol.
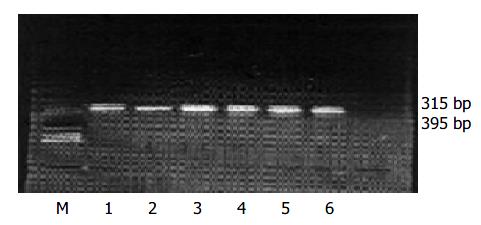
Twenty-eight samples were detected with both normalized PCR method and ELISA method. In the 14 samples positive for HBsAg, the results of the six antibody-positive amplifications are shown in Figure 1. The concordance of PCR with ELISA was 78.8%. Eight anti-HCV positive samples detected with multiplex PCR had a coincidence rate of 75% compared with ELISA method. When it was used to detect samples positive for both HBsAg and anti-HCV, the coincidence rate was 83.3%. Ten control cases negative for both HBsAg and anti-HCV were also detected with normalized PCR, of which 2 were positive for HBV and 1 for HCV.
Fourteen patients with HBV infection were detected with normalized PCR method. The results showed that the true positivity was 11 and the false negativity was 3 (Table 3).
| HBV patient | Non-HBV control | Total | |
| Positive | 11 | 2 | 13 |
| Negative | 3 | 8 | 11 |
| Total | 14 | 10 | 24 |
Of the eight HCV patients detected with normalized method, 6 were positive and 2 were negative. Six patients with both HBV and HCV infection were also detected, of which 5 were positive and 1 was negative (Figure 1 and Table 4). Ten patients without HBsAg and anti-HCV infection were also detected with our method, of which 8 were negative for HBV and 2 were positive for HBV (false positivity), 9 were negative for HCV and 1 positive for HCV (false positivity) (Table 3, Table 4).
| HCV patients | Non-HCV control | Total | |
| Positive | 6 | 1 | 7 |
| Negative | 2 | 9 | 11 |
| Total | 8 | 10 | 18 |
The sensitivity of this method for the detection of HBV DNA was 78.6% (11/14). The specificity was 80% (8/10). The diagnostic index was 158.6% (78.6% plus 80%) and the agreement rate was 79.2%. The positive predictive value was 84.6% (11/13). The negative predictive value was 72.7% (8/11) (Table 3).
The sensitivity of this method for the detection of HCV RNA was 75% (6/8). The specificity was 90% (9/10). The diagnostic index was 165% (75% plus 90%) and the diagnostic efficiency was 83.3%. The positive predictive value was 85.7% (6/7). The negative predictive value was 81.8% (9/11) (Table 4).
The sensitivity of this method for the detection of superinfection of HBV DNA and HCV RNA was 83.3% (5/6). The specificity was 70% (7/10). The diagnostic index was 153.3% (83.3% plus 70%) and the diagnostic efficiency was 72.2%. The positive predictive value was 62.5%. The negative predictive value was 87.5% (Table 5).
| HBV(+)and anti-HCV(+) | HBV(-) and anti-HCV(-) | Total | |
| Positive | 5 | 8 | 13 |
| Negative | 1 | 2 | 3 |
| Total | 6 | 10 | 16 |
Virus particles of HBV and HCV are rare in blood and HCV RNA is easily to degrade, therefore selecting protein denaturing agents to quickly dissolve the membrane protein is very important in the process of simultaneous extraction of HBV and HCV nucleic acids. Our results showed that method 4 was the most effective among the four extraction methods. The effective extraction rate was 95% for both HBsAg and anti-HCV positive serum. In method 4, hydrochloride guanine was a potent protein denaturant, it could dissolve the protein and destruct its secondary structure and cell structure, and mad it possible to separate nucleotide protein from nucleic acids. Urea and Triton X-100 are non-detergents and could dissolve cell membrane and precipitate the protein. Proteinase K could further degrade protein. Glass powder had absorptive effect on nucleic acids and therefore could absorb nucleic acids on its surface. Isopropyl alcohol could precipitate nucleic acids and finally the low ion potential solution could elute nucleic acids from the glass powder. This method was confirmed to be effective and authentic, suitable for the separation of virus RNA and DNA. In practice, it was a simple, fast, stable and reproducible method.
Theoretically, there is no limit on the number of target sequences simultaneously amplified by multiplex PCR. But the stringency of specific conditions restricted the number of target sequences amplified. In order to overcome these limits, we established a multiplex normalized amplification method. First a nested PCR amplification was performed with 2 pairs of primers with a joint sequence. Then a normalized amplification was done with joint primers. The effects of competition of primers were the lowest, so that the multiplex PCR became a single target PCR. These measurements could overcome the difficulties in establishing and optimizing the conditions of multiplex PCR. The success of this method has shed some lights on the development of PCR techniques. We used the orthogonal method for the design of experiment[6], and divided the key factors of PCR reaction such as primer concentration, dNTPs, Taq polymerase, AMV and magnesium chloride concentration into different grades, and a factor table was set up. This included all possible arrangements and combination of factors. The results showed that the coordination of proportion of the 6 factors was very important. If the proportion of HBV and HCV primers was not appropriate, and sometimes one of the amplifications was superior to another and only one band could be identified by electrophoresis.
ELISA is a method to detect antibody for the purpose of diagnosis of virus infection. The antibody was only detected 1 to 2 wk after infection, which reflected the immune response of the host, but could not explain the virus replication. PCR method could directly detect the virus nucleic acids. It could reflect the state of virus replication. When the virus was cleaned up, only the antibody was positive, the nucleic acids could not be detected. That is why the detection rate of PCR was lower when ELISA was used as a golden standard. In this study, 1/8 of the anti-HCV patients were negative for HCV RNA, and 1/ 6 patients were positive for both HBsAg and HCV, which were undetectable by our method. This is partly because the patients were in the state of convalescence, and the virus was already cleaned up or was false positive for ELISA due to hyperimmu-noglobulinemia, rheumatoid factor and superoxide dismutase. The detection of HCV RNA in non-hepatitis patients could be explained by the fact that the patients might be in the early stage of acute hepatitis, and the antibody had not been produced yet. A close follow-up is needed for the early diagnosis and treatment after the possibility of false positive is excluded.
Currently, the super-infection rate of HBV and HCV was very high, and it was 13.64% to 27.27% reported by Xu[7]. It is important to establish methods that can simultaneously detect combined virus infection. Konomi et al[8] reported a multiplex polymerase chain reaction (PCR) method for simultaneous detection of hepatitis B, C, and G viral genomes. The levels of concordance with the data obtained by conventional single PCR method were 100% for single infection, 98-100% for double infection, and 92% for triple infection. Meng Q and his colleagues[9] established an automatic multiplex system for simultaneously screening hepatitis B virus (HBV), hepatitis C virus (HCV), and human immunodeficiency virus type 1 (HIV-1) in blood donations. The detection limits (95% confidence interval) were 22 to 60 copies/mL for HBV, 61 to 112 IU/mL for HCV, and 33 to 66 copies/mL for HIV-1, using a specimen input volume of 0.2 mL. The AMPLINAT MPX assay could detect a broad range of genotypes or subtypes for all three viruses and had a specificity of 99.6% for all three viruses with sero-negative specimens. In an evaluation of sero-conversion panels, the AMPLINAT MPX assay detected HBV infection an average of 24 d before the detection of HBsAg by enzyme immunoassay. HCV RNA was detected an average of 31 d before HCV antibody appeared. HIV-1 RNA was detected an average of 14 d before HIV-1 antibody and an average of 9 d before p24 antigen. A Chinese group has designed a visual gene-detecting technique using nanoparticle-supported gene probes. With the aid of gold nanoparticle-supported 3’-end-mercapto-derivatized oligonucleotide serving as a detection probe, and 5’-end - amino-derivatized oligonucleotide immobilized on glass surface acting as a capturing probe, target DNA was detected visually by sandwich hybridization based on highly sensitive “nano-amplification” and silver staining. Different genotypes of hepatitis B and C viruses in serum samples from infected patients were detected using home made HBV, HCV, and HBV/HCV gene chips by the gold/silver nanoparticle staining amplification method. The present visual gene-detecting technique might avoid limitations of the reported methods due to for its high sensitivity, good specificity, simplicity, speed, and cheapness[10].
Multiplex PCR assay has been used for the simultaneous detection of many different genes of pathogens including genes related with antibiotic resistance genes in Staphylococcus aureus[11]. A multiplex reverse transcriptase polymerase chain reaction (RT-PCR) was also applied for the simultaneous detection of hepatitis A virus (HAV), poliovirus (PV) and simian rotavirus (RV-SA11), and compared with specific primers for each genome sequence. Three amplified DNA products representing HAV (192 bp), PV (394 bp) and RV (278 bp) were identified when positive controls were used[12]. A multiplex RT-PCR method was described for the simultaneous detection of all four viruses in combination with a plant mRNA specific internal control which could be used as an indicator of the effectiveness of the extraction and RT- PCR. The upper detection limit for the four viruses was at an extract dilution of 1/200[13]. A multiplex semi-nested PCR was developed for the simultaneous detection and differentiation among porcine circovirus 1 (PCV1), PCV2, and porcine parvovirus (PPV) from boar semen. Primers of PCV1, PCV2 and PPV were specific and did not react with other viruses respectively. Twenty (20.4%) and 42 (42.9%) out of 98 whole semen samples were found to be positive for PCV and PPV using conventional multiplex and semi-nested PCR respectively[14]. Multiplex method for HBV/HCV/HIV-1 has been used for screening 6 805 010 units of serologically negative donation and 112 HBV DNA-positives, 25 HCV RNA positives and 4 HIV-1 RNA positives were screened out and prevented transfusion of the positive blood[15,16].
The sensitivity of our multiplex normalized PCR method was 78.6%, 75% and 83.3% for the detection of HBVDNA, HCVRNA, and super-infection of HBV and HCV respectively. The specificity was 80%, 90% and 70%, respectively. These are good enough for a diagnostic assay. It can detect both DNA and RNA simultaneously and can be completed in one day. It is not only suitable for clinical diagnosis, but also suitable for the screening of HBV and HCV from blood donators to prevent the transmission of these diseases. It can also be used for an epidemiological study. In these respects it needs to be further studied in a large-scale population.
Edited by Wang XL and Zhang JZ Proofread by Xu FM
| 1. | Hu KQ, Yu CH, Lee S, Villamil FG, Vierling JM. Simultaneous detection of both hepatitis B virus DNA and hepatitis C virus RNA using a combined one-step polymerase chain reaction technique. Hepatology. 1995;21:901-907. [PubMed] |
| 2. | Yamada O, Matsumoto T, Nakashima M, Hagari S, Kamahora T, Ueyama H, Kishi Y, Uemura H, Kurimura T. A new method for extracting DNA or RNA for polymerase chain reaction. J Virol Methods. 1990;27:203-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 3. | Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495-503. [PubMed] |
| 4. | Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979;76:615-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 910] [Cited by in RCA: 1064] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 5. | Han JX, Zhang C, Yang XC, Tang TH, Wang ML. Investigation on the purification and synthesizing of DNA fragments by OPC methods. Shandong Yike Daxue Xuebao. 1995;33:173-174. |
| 6. | Fang JQ, Fu CZ, Liao RR. Regression methods of pair -wise data. Zhongguo Weisheng Tongji. 1996;13:1-5. |
| 7. | Xu ZF, Xu KC, Meng XY. Investigation on the double infection of HBV and HCV. Nantong Yixueyuan Xuebao. 1994;14:162-165. |
| 8. | Konomi N, Yamaguchi M, Naito H, Aiba N, Saito T, Arakawa Y, Abe K. Simultaneous detection of hepatitis B, C, and G viral genomes by multiplex PCR method. Jpn J Infect Dis. 2000;53:70-72. [PubMed] |
| 9. | Meng Q, Wong C, Rangachari A, Tamatsukuri S, Sasaki M, Fiss E, Cheng L, Ramankutty T, Clarke D, Yawata H. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2001;39:2937-2945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Wang YF, Pang DW, Zhang ZL, Zheng HZ, Cao JP, Shen JT. Visual gene diagnosis of HBV and HCV based on nanoparticle probe amplification and silver staining enhancement. J Med Virol. 2003;70:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 46] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089-4094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 391] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 12. | Coelho C, Vinatea CE, Heinert AP, Simões CM, Barardi CR. Comparison between specific and multiplex reverse transcription-polymerase chain reaction for detection of hepatitis A virus, poliovirus and rotavirus in experimentally seeded oysters. Mem Inst Oswaldo Cruz. 2003;98:465-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 13. | Thompson JR, Wetzel S, Klerks MM, Vasková D, Schoen CD, Spak J, Jelkmann W. Multiplex RT-PCR detection of four aphid-borne strawberry viruses in Fragaria spp. in combination with a plant mRNA specific internal control. J Virol Methods. 2003;111:85-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Kim J, Han DU, Choi C, Chae C. Simultaneous detection and differentiation between porcine circovirus and porcine parvovirus in boar semen by multiplex seminested polymerase chain reaction. J Vet Med Sci. 2003;65:741-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Ohnuma H, Tanaka T, Yoshikawa A, Murokawa H, Minegishi K, Yamanaka R, Lizuka HY, Miyamoto M, Satoh S, Nakahira S. The first large-scale nucleic acid amplification testing (NAT) of donated blood using multiplex reagent for simultaneous detection of HBV, HCV, and HIV-1 and significance of NAT for HBV. Microbiol Immunol. 2001;45:667-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Mine H, Emura H, Miyamoto M, Tomono T, Minegishi K, Murokawa H, Yamanaka R, Yoshikawa A, Nishioka K. High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J Virol Methods. 2003;112:145-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |