Published online Jun 1, 2004. doi: 10.3748/wjg.v10.i11.1666
Revised: January 4, 2003
Accepted: February 17, 2003
Published online: June 1, 2004
AIM: To establish a method for optical sections of HepG2 human hepatoblastoma cells with confocal laser scanning microscope (CLSM) and to study the spatial structure of filamentous actin (F-actin) in HepG2 cells.
METHODS: HepG2 cells were stained with FITC-phalloidin that specifically binds F-actin, with propidium iodide (PI) to the nucleus, and scanned with a CLSM to generate optically sectioned images. A series of optical sections taken successively at different focal levels in steps of 0.7 mm were reconstructed with the CLSM reconstruction program.
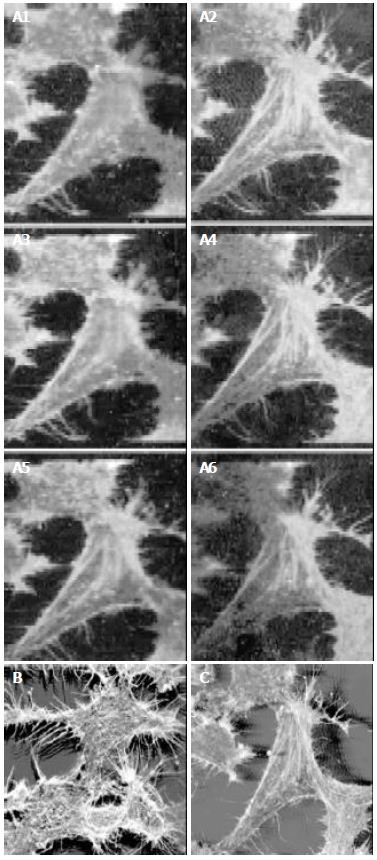
RESULTS: CLSM images showed that the FITC-stained F-actin was abundant microfilament bundles parallel or netted through the whole cell and its processes. Most F-actin microfilaments extended through the cell from one part toward the other or run through the process. Some microfilaments were attached to the plasma membrane, or formed a structural bridge connecting to the neighboring cells.
CONCLUSION: A method for double labeling HepG2 human hepatoblastoma cells and CLSM imaging F-actin microfilaments and nuclei by image thin optical sections and spatial structure was developed. It provides a very useful way to study the spatial structure of F-actin.
- Citation: Huo X, Xu XJ, Chen YW, Yang HW, Piao ZX. Filamentous-actins in human hepatocarcinoma cells with CLSM. World J Gastroenterol 2004; 10(11): 1666-1668
- URL: https://www.wjgnet.com/1007-9327/full/v10/i11/1666.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i11.1666
Cytoskeleton, consisting of proteins, is the structural network in cells. It closely relates to the functions of cells, such as cell movement, cell morphology and transmenbrane signal conduction, etc. Observation and analysis of cell F-actin with CLSM are a precise, rapid, and simple method. This paper aimed at using CLSM in combination with double label for the observation and analysing of the three-dimensional structure of cytoskeleton system in HepG2 hepatocarcinoma cells.
HepG2 cells were inoculated in improved Petri dish (normal Petri dishes with a center hole bottomed by quartz glass), with 100 mL/L BAS/RPMI-1640 as culture medium for 24-48 h in an incubator at 37 °C with a 950 mL/L air/50 mL/L CO2 atmosphere. After the dish glass was covered fully with cells, the culture medium was poured out. The cells were rinsed with PBS twice, and then fixed in 40 g/L paraformaldehyde for 30 min at 4 °C.
The above-mentioned specimens were rinsed with PBS twice, then permeabilized for 10 min in 2 g/L Triton X-100 at room temperature, and nonspecific background was blocked using 5 g/L BSA for a while. Specimens were then stained with 5 mg/L FITC-phalloidin (Sigma) at room temperature for 30 min. After that, the specimens were rinsed with PBS twice and stained with 5 mg/L PI (Sigma), and finally rinsed twice with PBS and a little PBS in dishes was reserved for observation. All procedures must be performed from light.
Specimens were examined with an ACAS Ultima 312 CLSM (Meridian Instruments, USA) equipped with an argon ion laser providing simultaneous excitation at 514.5 nm, 488 nm and 351-364 nm, an output power of 50 mW for the UV and 200 mW for the combined visible wavelengths, a high-precision mechanical scanning stage (0.1 mm in the X-Y plane and 0.1 mm in the Z axis), and an inverted microscope (Olympus), three photomultiplier tubes (PMT). The availability of a variety of excitation wavelengths on a single microscope setup offers the power and convenience of using combinations of fluorescent probes in a single specimen. In this experiment, the parameters were as follows: Objective: 100 × 1.30 oil, Pinhole: 255 mm, X points: 270, Y point: 290, Z step: 0.7 mm, Data model: Z image, Scan Type: Mirror, Step Size: 0.20 mm, Speed: 20 mm/s, Samples/pt: 15, Laser Power: 612 mw, Scan stress: 100%, PMT 1: 40%, PMT 2: 40%, Z point: 15. According to the above parameters, a series of images were collected sequentially from single optical sections at 0.7 mm intervals (15 planes per series) using the 488, 530 nm laser lines to excite FITC and PI, respectively. Randomly selected regions of specimens were scanned to image using Z-series program of CLSM. With the program the specimens could be scanned cross-sectionally along the X, Y, and Z axes, and serial optical sections of different layers could be obtained without injury. Firstly, the cell top was confirmed by prescanning, and then the specimen from top to bottom was scanned according to Z-step to obtain optical sections layer by layer, and each image was recorded. The images were electronically colored. The optical sections were reconstructed to stereoscopic images with CLSM reconstruction program.
The HepG2 cells stained with FITC-phalloidin and PI were successfully labeled. There were six serial optical sections of HepG2 cells as shown in Figure 1. The interval between two sections was fixed at 0.7 mm. The images shown on Figure 1a2 - Figure 1a4 were all three-dimensional images obtained from sequential optical sections, which were reconstructed with the reconstruction program of CLSM. The HepG2 cells, having many primary and secondary processes, were irregular in shape. The FITC-stained F-actin filaments appeared in bundles, which ran in parallel to the main cell axis or through cell processes. Some microfilaments were attached to the plasma membrane, or formed a structural bridge connecting to the neighboring cells. When detector 1 of CLSM for 488 nm line was on, only F-actin marked with FITC-phalloidin could be shown (Figure 1). When both of the two detectors of CLSM were turned on, F-actin marked with FITC-phalloidin and the nuclei marked with PI could be shown simultaneously (Figure 2). F-actin appeared green, the distinct nuclei appeared red, and the region around the nuclei was overlapped by green F-actin and red nuclei appeared yellow.
Cytoskeleton, composed largely of actin filaments, is the internal framework of a cell. It includes micro-tubes, microfilaments and middling fibers, and is the base structure for cell movement, cell morphology and transmembrane signal conduction. Microfilament mainly consists of actin participating in keeping cell pattern and tight junction between cells. It is also related to the adhesion of the ground substance outside the cells. Microfilament exists in the form of G-actin when it is dissociated or exists in the form of F-actin when it is polymerized. The change and balance of microfilament dissociation and polymerization in normal cells are the important regulatory factors for the movement, adhesion and fission cycle of cells[1,2]. Cell mutation involves cytoskeletal rearrangement and morphological alterations. As a sensitive norm in examining the development of tumor cells at the early stage, F-actin has been used in human mammary gland cancer and T-lymphocyte cancer[3-5]. Our result showed that F-actin microfilaments of HepG2 cells existed in the form of bunches of fibers that were thick, long and dense. F-actin filaments formed bundles running through the main cell axis or cellular processes. The relationship between the change of microfilament morphology and cancer occurrence in cells is still unknown. But when microfilament’s component and distribution change, cells cannot receive normal regulation. And it leads to the loss of control of cell growth and propagation and to cancer occurrence. According to some researches, the change of microfilaments might reduce the gap junction in number and function, and the communication function of cells, which is the main reason why propagation ability of carcinoma cells rose up and the ability of adherence lowered, and easily fell off[6-9]. The mechanism underlying the morphological changes of microfilaments in the process of cancer cells is unclear.
In this experiment, F-actin was marked with FITC-phalliodin and the nuclei with PI, so the cytoskeleton and nuclei could be observed. This provides a very useful way to study the relationship between cytoskeleton and nucleus. Compared with other methods, such as immunohistochemistry, light microscopy or electrical microscopy[10-12], CLSM has several advantages[13-17]: Firstly, resolution obtained with a CLSM is better than that of conventional microscopy because of the combination of laser illuminator and detection pinhole. With conventional light microscopy, the fluorescence of the entire specimen interferes with the resolution, while CLSM allows examining the organization of fluorescent labeled cells and tissues by eliminating the “out-of-focus” flare of the specimen, thus providing sharp, high-contrast images of cells and subcellular structures within thick samples. Since images are obtained by scanning, excessive illumination of the specimen and quick decrease of the fluorescent signal are avoided. And light bleaching is minimized because exposure to light is restricted to the area of the preparation being examined. Secondly, this method of embedding and sectioning specimens, which are a time-consuming and a destructive procedure, can now be avoided. So it is quick and simple to make samples using CLSM. Under conventional microscopy, structures present in the tissues can be observed only after sectioning (microtome sections). With the CLSM it is possible to obtain a number of serial optical sections of quite thick specimens without physical sectioning. Furthermore, serial optical sections can be collected and displayed as digitalized images, and by using a computer, the digitalized images obtained may be reconstructed as a 3-dimensional structure. In addition, the procedure is not destructive for a living cell or tissue. In this experiment using CLSM, the images of the cytoskeleton marked by the double label technique and the stereoscopy of three-dimensional images were very clear. As the sectioning with CLSM is not done physically but optically, the procedure is not destructive for an intact cell, and the process of sectioning turns simpler and quicker and more reliable, and it can notably keep the normal morphology and function of the cell.
In summary, our results showed that in HepG2 cells F-actin microfilaments existed in the form of bunches of fibers that were thick, long and dense, and ran through the main cell axis or cellular processes. It is suggested that F-actin may play an important role in HepG2 cells.
| 2. | Shumilina EV, Negulyaev YA, Morachevskaya EA, Hinssen H, Khaitlina SY. Regulation of sodium channel activity by capping of actin filaments. Mol Biol Cell. 2003;14:1709-1716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 3. | Sugiura T, Nakane S, Kishimoto S, Waku K, Yoshioka Y, Tokumura A. Lysophosphatidic acid, a growth factor-like lipid, in the saliva. J Lipid Res. 2002;43:2049-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 4. | Shestakova EA, Wyckoff J, Jones J, Singer RH, Condeelis J. Correlation of beta-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 1999;59:1202-1205. [PubMed] |
| 5. | Williams JI, Weitman S, Gonzalez CM, Jundt CH, Marty J, Stringer SD, Holroyd KJ, Mclane MP, Chen Q, Zasloff M. Squalamine treatment of human tumors in nu/nu mice enhances platinum-based chemotherapies. Clin Cancer Res. 2001;7:724-733. [PubMed] |
| 6. | Autieri MV, Carbone C, Mu A. Expression of allograft inflammatory factor-1 is a marker of activated human vascular smooth muscle cells and arterial injury. Arterioscler Thromb Vasc Biol. 2000;20:1737-1744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 7. | Yamakita Y, Oosawa F, Yamashiro S, Matsumura F. Caldesmon inhibits Arp2/3-mediated actin nucleation. J Biol Chem. 2003;278:17937-17944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 8. | Yan H, Rivkees SA. Hepatocyte growth factor stimulates the proliferation and migration of oligodendrocyte precursor cells. J Neurosci Res. 2002;69:597-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J Cell Biol. 2003;161:371-380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Uchida M, Hanai S, Uematsu N, Sawamoto K, Okano H, Miwa M, Uchida K. Overexpression of poly (ADP-ribose) polymerase disrupts organization of cytoskeletal F-actin and tissue polarity in Drosophila. J Biol Chem. 2002;277:6696-6702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer's disease pathophysiology. FASEB J. 2001;15:2433-2444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 496] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 12. | Fischer RS, Lee A, Fowler VM. Tropomodulin and tropomyosin mediate lens cell actin cytoskeleton reorganization in vitro. Invest Ophthalmol Vis Sci. 2000;41:166-174. [PubMed] |
| 13. | Masuda T, Fujimaki N, Ozawa E, Ishikawa H. Confocal laser microscopy of dystrophin localization in guinea pig skeletal muscle fibers. J Cell Biol. 1992;119:543-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Huo X, Lu JX, Yang RD, Li ZZ, Wei WX, Zheng XH, Tu WX, Xiao ZX, Zhang FL. Comparison of laser scanning confocal microscope with light microscope. Jiguang Shengwu Xuebao. 2001;10:76-79. |
| 15. | Dailey M, Marrs G, Satz J, Waite M. Concepts in imaging and microscopy. Exploring biological structure and function with confocal microscopy. Biol Bull. 1999;197:115-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Li J, Jester JV, Cavanagh HD, Black TD, Petroll WM. On-line 3-dimensional confocal imaging in vivo. Invest Ophthalmol Vis Sci. 2000;41:2945-2953. [PubMed] |
| 17. | Benbow U, Orndorff KA, Brinckerhoff CE, Givan AL. Confocal assay for invasion: use of propidium iodide fluorescence and laser reflectance to quantify the rate of migration of cells through a matrix. Cytometry. 2000;40:253-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |