Published online Jun 1, 2004. doi: 10.3748/wjg.v10.i11.1565
Revised: August 4, 2003
Accepted: August 16, 2003
Published online: June 1, 2004
AIM: To investigate cytpchrome P4502E1 (CYP2E1) gene expression in occurrence and progression of hepatocellular carcinoma (HCC).
METHODS: The human liver arrayed library was spotted onto the nylon membranes to make cDNA array. Hybridization of cDNA array was performed with labeled probes synthesized from RNA isolated from HCC and adjacent liver tissues. Sprague-Dawley rats were administrated diethylnitrosamine (DENA) to induce HCC. CYP2E1 expression was detected by the method of RT-PCR and Northern blot analysis.
RESULTS: CYP2E1 was found by cDNA array hybridization to express differently between HCC and liver tissues. CYP2E1 only expressed in liver, but did not express in HCC tissues and expressed lowly in cirrhotic tissues. In the progression of cirrhosis and HCC, the expression level of CYP2E1 was gradually decreased and hardly detected until the late stage of HCC.
CONCLUSION: Using arrayed library to make cDNA arrays is an effective method to find differential expression genes. CYP2E1 is a unique gene expressing in liver but did not express in HCC. CYP2E1 expression descended along with the initiation and progression of HCC, which is noteworthy further investigations in its significance in the development of HCC.
-
Citation: Man XB, Tang L, Qiu XH, Yang LQ, Cao HF, Wu MC, Wang HY. Expression of
cytochrome P4502E1 gene in hepatocellular carcinoma. World J Gastroenterol 2004; 10(11): 1565-1568 - URL: https://www.wjgnet.com/1007-9327/full/v10/i11/1565.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i11.1565
Hepatocellular carcinoma (HCC) is one of the most common cancers in China and the world[1,2]. Although the wide use of diagnostic technology and the improvement in curative treatment may evolve to a better scenario, it still represents more than 5% of all cancers[3]. To investigate HCC associated genes is very helpful to elucidating the molecular mechanism of proliferation, differentiation and transformation of hepatocytes in the occurrence and development of HCC[4,5]. cDNA microarray analysis is a powerful technique in the investigation of cancer associated gene identification and function[6]. The gene expression can be simultaneously monitored in a large scale with cDNA microarray[7]. The potential analysis of the expression of thousands of genes in one experiment provided new insights into the molecular study of the occurrence and development of HCC[8-10].
In the present study, a method of making cDNA array from the arrayed library was developed to identify the differentially expressed genes. CYP2E1, the gene encoding cytochrome P450 2E1 (CYP2E1), a member of cytochrome P450s present in prokaryotes and through the eukaryotes[11], was identified to express in normal liver or cirrhotic tissues adjacent to tumors but not express in HCC tissues. CYP2E1 is one of the important members of cytochrome P450 superfamily, with functions ranging from catalysis of the conversion of ethanol to acetaldehyde and from acetate to metabolization of many exotic drugs and procarcinogens[12]. A rat HCC model was then induced to study CYP2E1 expression in the procession of HCC. The results showed that CYP2E1 expression descended along with the initiation, promotion and progression of HCC. It is suggested that CYP2E1 is correlated to HCC and noteworthy further investigations for its significance in the development of HCC.
The human liver cDNA library (Invitrogen, USA) was cultured on the agar plate and were picked into 96-well microplates with 200 mL culture medium. After an overnight culture, 1 mL of the bacterial medium in each well was diluted into 20 mL from which 1 mL was transferred to the corresponding 96-well PCR microplates and the remaining was added to 50 mL glycerol and stored at -80 °C.
PCR reaction was carried out with oligonucleotide T7 (5’ gga aga agg gaa ctg att cag 3’) and oligonucleotide BGHR (5’ cac atc cag atc ata tgc cag 3’) as forward primer and reverse primer. The procedure of PCR was made with denaturing at 94 °C for 4 min followed by 35 cycles of reaction including denaturing at 94 °C for 50 s, annealing at 58 °C for 50 s and elongation at 72 °C for 90 s, and a final bonus extension elongation at 72 °C for 7 min. The amplified products were randomly selected for electrophoresis to validate PCR efficiency.
The PCR products in the microplates were spotted with TAS (BioRobotics, UK) onto the 8 cm × 12 cm nylon membrane to form 2 × 2 × 96 array in each membrane. The 0.7 mm diameter 96-pin spotting setting was used. Each product from a well was spotted onto the same position 3 times. The membranes were denatured immediately in the denature buffer (1.5 mol/L NaCl, 0.5 mol/L NaOH) for 5 min and then equalized in the equalizing buffer (0.9 mol/L NaCl, 0.5 mol/L Tris, pH7.5) for 5 min followed by baking at 80 °C.
DENA (Sigma, USA) was diluted into 1 × 10-4 concentration in drinkable water. Male Sprague-Dawley rats were obtained from the Experimental Animal Center of Chinese Academy of Sciences, Shanghai. The rats in HCC group were administrated DENA via drinking DENA-diluted water while the rats in control group drank clean water. Three HCC-induced rats and one control rat were sacrificed by decollation under pentobarbital anesthesia every week. The liver tissue was immediately stored in liquid nitrogen for RNA isolation and fixed for histological analysis.
Total RNA was isolated from 0.1 g frozen tissues in 1 mL Trizol™ reagent (Invitrogen, USA) according to the manufacturer’s instructions. Isolation of mRNA was carried out with the Oligotex™ mRNA Mini kit (Qiagen, Germany) from 250 mg total RNA.
Probes for the array hybridization were labeled with Atlas™ SpotLight™ labeling kit (Clontech, USA) according to the user’s manual. A 2 mg mRNA respectively from paired HCC tumor and adjacent normal or cirrhotic tissues was added with 2.5 mL CDS Primer Mix and incubated at 70 °C for 10 min. Reaction buffer (5 ×) 5 mL, Labeling Mix (10 ×) 2.5 mL, DTT (100 mmol/L) 1.25 mL were added and incubated at 48 °C for 5 min. PowerScript reverse transcriptase (1.25 mL) per reaction and 10 mL of Master Mix were added and incubated at 48 °C for an additional 45 min. The reaction was stopped by adding 0.5 mL of 0.5 mol/L EDTA (pH8.0) and then purified routinely.
DNA array hybridization and detection were preceded with SpotLight™ chemiluminescent hybridization & detection kit (Clontech, USA). The array membranes were wetted and pre-hybridized while a biotinylated array probe was denatured. Cot-1 and the biotinylated probe were added to pre-hybridization solution to incubate overnight. The membranes were washed and blocked, and added with enough stabilized Streptavidin-HRP conjugate. After equilibrated, the membranes were covered with the luminol/peroxide working solution and incubated at room temperature for 5 min and exposed the membrane to film for an appropriate time to obtain desired signals.
First strand cDNA was reversibly transcribed from total RNA with SuperScript™ II reverse transcriptase (Invitrogen, USA). The procedure of RT-PCR of CYP2E1 was carried out in normal rat tissues with the oligonucleotide (5’ act tct acc tgc tga gca c 3’) and oligonucleotide (5’ ttc agg tct cat gaa cgg g 3’) as forward and reverse primer respectively and with denaturing at 94 °C for 4 min followed by 33 cycles of reaction including denaturing at 94 °C for 50 s, annealing at 55 °C for 50 s and extension at 72 °C for 1 min, and a final bonus extension for 7 min. An 874-bp sequence product was amplified.
The PCR product was purified using the QIAquick PCR purification kit (Qiagen, Germany) and cloned into the T-vector (Promega, USA) directly and transfected into DH5-alpha bacteria. The plasmid was purified to be sequenced and confirmed according to the GenBank sequence.
The plasmid was digested and electrophoresed. The template fragment was purified from agarose gel using the gel extracting kit (Qiagen, Germany). The probe was labeled from 25 ng template DNA by the random primer method using DNA polymerase I large (Klenow) fragment (Promega, USA) with a-32p-dCTP followed by purification using the QIAquick purification kit (Qiagen, Germany). Total RNA (40 mg) justified by 28 s and 18 s intensity of each sample, was loaded. Electrophoresis was carried out under denaturing conditions and RNA was transferred onto nitrocellulose membrane and cross-linked by baking at 80 °C for 2 h. The filters were then prehybridized, hybridized, and washed under high stringency conditions. All blots were screened by Fuji BAS2000 and analyzed by LABwork software and then exposed at -80 °C to Kodak X-ray film for 2 wk.
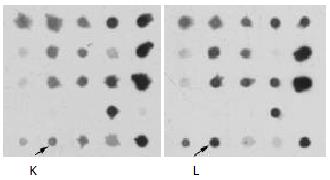
The arrayed library clones were spotted onto each nylon membrane to 2 × 2 × 96 array from 4 microplates. There were 96 subarrays in one membrane. The arrayed membranes were denatured, fixed and preserved under dry condition. The mRNA from HCC tissues and corresponding adjacent liver tissues was labeled to hybridize to the above arrayed membranes. A spot then identified low signals in HCC tissues but high signals in corresponding adjacent liver tissues (Figure 1). It was suggested that the gene was a liver-expressing gene that was downregulated in HCC tissues. According to the position of the spot in membrane, the clones in microplates of arrayed library was recruited and sequenced as CYP2E1.
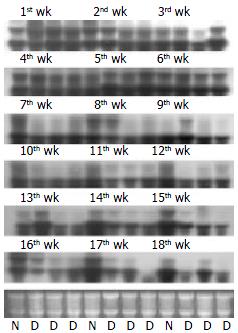
The liver tissue from DENA-induced rats that were killed after 3 wk appeared almost normal as the control rats. The pathological examination indicated the liver lesion in these rats. The livers of rats after the 4th wk were smaller than those of control rats and the color of the liver surface was much dingy. Cirrhotic nodi could be seen on the liver surface of rats killed in the 7th wk and cirrhosis could be detected as early as in the 5th wk by pathological examination. There were few liver cancer nodi in 1 rat detected in the 9th wk. So the 9th wk might be the boundary between cirrhosis and HCC in the present study. In the 10th wk, HCC could be detected in all rats and HCC nodi could be seen by naked eyes after the 10th wk. After the 16th wk, HCC nodi were spread almost all over the rat liver.
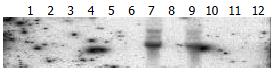
Twelve types of human normal tissues except liver were used for Northern blot analysis of CYP2E1 expression. CYP2E1 did not express at all in the 12 normal human tissues. To represent the expression level of CYP2E1 in these organs, the RNA sample of heart, brain, lung, intestine and liver were selected with 2 cases of HCC tissues and 1 case of kidney cancer tissue to be transferred onto one membrane for Northern blot analysis. The hybridization result showed that CYP2E1 indeed only expressed in non-tumor liver tissues (Figure 2). The expression pattern of CYP2E1 in rats was the same as in human. By RT-PCR analysis, it was indicated that CYP2E1 was only amplified from normal liver tissues and did not express in other tissues (Figure 3).
Northern blot analysis showed that CYP2E1 expressed in normal liver tissues and adjacent tissues with cirrhosis. There was no hybridization signal in HCC tissues from all 14 cases. CYP2E1 was significantly downregulated in HCC (Figure 4). The rat HCC model simulated the progression of HCC. The change of expression was analyzed also by Northern blot (Figure 5). In the early liver lesion stage of the DENA-induced rats after 1 - 3 wk, CYP2E1 expression was slightly upregulated compared with normal livers. But after 4 wk, the gene was down-regulated and when cirrhosis occurred after 5 wk, the expression level was almost the same in the liver between the 2 groups. It was obvious that the cirrhotic tissues of the rats expressed at lower level than the normal livers after 7 wk. When the DENA-induced rats progressed to the stage of tumor from the 10th wk, CYP2E1 expressed at much lower level than normal livers and liver of cirrhotic stages. Until the very later stage of HCC, the expression level was very low and the hybridization signal was hardly detected.
An ideal support allowed effective immobilization of probe onto its surface, and robust hybridization of target with the probe[13]. Nylon membrane, as a standard support used for making microarrays[14,15] could hold more DNA. The hybridization and detection of membranes arrays cost much less. Here we used the nylon membranes as the matrix of cDNA arrays. The DNA on the membranes was denatured before cross-linking to the matrix by baking the array at 80 °C. In many cases, the cDNA targets were chosen directly from cDNAs library of interest for DNA microarray manufacturing[16]. The collection should include an aggregation of gene clones as many as possible. On the other hand, for certain purpose of an investigation, most genes on a large content array might not be necessary. The method to make cDNA microarray in this study arose from the concept of arrayed library. The cDNA plasmid library was transferred to the microplates to be the arrayed library, which was copied to the nylon membranes to make the DNA arrays after PCR amplification. Because most genes on the arrays did not differently express between samples, the tumor and normal tissues, for example, the sequencing expense of these genes could be saved. It was after the array hybridization, the genes that were detected to express differently would be selected from the arrayed library and were sequenced for further studies. In this study, most genes showed no differential expression between HCC and adjacent liver tissues. Only the differently expressed genes were sequenced for further investigations. CYP2E1 was identified as a liver profound but HCC silent gene.
P450 enzymes were found in almost all eukaryotes and prokaryotes[17]. CYP2E1 enzyme had a molecular weight of 57 KD and the encoding gene was located on chromosome 10 spanning 11413 base pairs[18,19]. CYP2E1 was found to play an important role in its polymorphism during tumor occurrence. The significance of this polymorphism currently unclear[20,21]. To date, the results in possible associations between CYP2E1 genetic polymorphisms and alcoholic liver disease susceptibility have been varied and often contradictory[22]. Because hepatic CYP2E1 has been shown to activate various carcinogens, there has been interest in whether certain CYP2E1 polymorphisms might predispose to liver cancer[23,24]. It was demonstrated that possession of the less common Rsa I/Pst I allele was associated with increased susceptibility to HCC[25].
Although CYP2E1 was said to be located in most tissues with the largest concentration in the liver, it was constitutively expressed in the liver and only induced to express in other tissues by treatment with acetone, ethanol, isoniazid and other compounds, many of which are substrates for the enzyme[26-28]. This study showed that CYP2E1 expressed only in the liver but did not express in the other normal organs in human and rats. So it could be concluded that CYP2E1 is the liver-specific functional gene. Furthermore, CYP2E1 was found to loose expression in HCC. Northern blot analysis was performed to validate the result of chip experiment. CYP2E1 expressed at a high level in normal or cirrhotic tissues but was silent in HCC tissues from 14 cases of HCC. So CYP2E1 did not express in HCC. To further understand the expression of CYP2E1 in the initiation, promotion and progression of HCC, the DENA-induced rat HCC model was made to study the role of CYP2E1 in the process of HCC. The development of rat HCC underwent the progression of liver lesion and cirrhosis until the tumor occurrence. The results showed that at the early stage of the rat model the expression level of CYP2E1 in the liver lesion tissues was slightly higher than that in normal rat liver in the first to third week. Because CYP2E1 was involved in the metabolism of nitrosamines, DENA might induce CYP2E1 expression. But along with the aggravation of liver lesion and development of cirrhosis, the expression level of CYP2E1 was gradually descended. After the progressing stage of HCC, CYP2E1 did not express as in human HCC. There were very low signals because at the late stages of HCC the tumor were grown sporadically and the samples were intermixed with cirrhotic tissues where CYP2E1 still expressed. The expression pattern of CYP2E1 in the HCC was worth of further studies. It is suggested that these differential expression might help further understand the molecular genetic and gene regulation mechanism of HCC progression. Because CYP2E1 does not express in HCC cells, there might be a guided biological treatment for HCC with its potential substrate.
Edited by Ma JY and Wang XL Proofread by Xu FM
| 1. | Qin LX, Tang ZY. The prognostic significance of clinical and pathological features in hepatocellular carcinoma. World J Gastroenterol. 2002;8:193-199. [PubMed] |
| 2. | Ince N, Wands JR. The increasing incidence of hepatocellular carcinoma. N Engl J Med. 1999;340:798-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 109] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 3. | Llovet JM, Beaugrand M. Hepatocellular carcinoma: present status and future prospects. J Hepatol. 2003;38 Suppl 1:S136-S149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 5. | Tannapfel A, Wittekind C. Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis. Virchows Arch. 2002;440:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Berns A. Gene expression in diagnosis. Nature. 2000;403:491-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Young RA. Biomedical discovery with DNA arrays. Cell. 2000;102:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 224] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929-1939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Delpuech O, Trabut JB, Carnot F, Feuillard J, Brechot C, Kremsdorf D. Identification, using cDNA macroarray analysis, of distinct gene expression profiles associated with pathological and virological features of hepatocellular carcinoma. Oncogene. 2002;21:2926-2937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Lee JS, Thorgeirsson SS. Functional and genomic implications of global gene expression profiles in cell lines from human hepatocellular cancer. Hepatology. 2002;35:1134-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Peterson JA, Graham SE. A close family resemblance: the importance of structure in understanding cytochromes P450. Structure. 1998;6:1079-1085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 84] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | Tanaka E, Terada M, Misawa S. Cytochrome P450 2E1: its clinical and toxicological role. J Clin Pharm Ther. 2000;25:165-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 143] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21:5-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 544] [Cited by in RCA: 469] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 14. | Schlaak JF, Hilkens CM, Costa-Pereira AP, Strobl B, Aberger F, Frischauf AM, Kerr IM. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J Biol Chem. 2002;277:49428-49437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. Identification of transforming growth factor-beta- regulated genes in caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci USA. 1999;96:15020-15025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999;21:15-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 359] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 17. | Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2078] [Cited by in RCA: 1912] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 18. | Umeno M, McBride OW, Yang CS, Gelboin HV, Gonzalez FJ. Human ethanol-inducible P450IIE1: complete gene sequence, promoter characterization, chromosome mapping, and cDNA-directed expression. Biochemistry. 1988;27:9006-9013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 132] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Umeno M, Song BJ, Kozak C, Gelboin HV, Gonzalez FJ. The rat P450IIE1 gene: complete intron and exon sequence, chromosome mapping, and correlation of developmental expression with specific 5' cytosine demethylation. J Biol Chem. 1988;263:4956-4962. [PubMed] |
| 20. | Tsutsumi M, Takada A, Wang JS. Genetic polymorphisms of cytochrome P4502E1 related to the development of alcoholic liver disease. Gastroenterology. 1994;107:1430-1435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 90] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Ueshima Y, Tsutsumi M, Takase S, Matsuda Y, Kawahara H. Acetaminophen metabolism in patients with different cytochrome P-4502E1 genotypes. Alcohol Clin Exp Res. 1996;20:25A-28A. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Wong NA, Rae F, Simpson KJ, Murray GD, Harrison DJ. Genetic polymorphisms of cytochrome p4502E1 and susceptibility to alcoholic liver disease and hepatocellular carcinoma in a white population: a study and literature review, including meta-analysis. Mol Pathol. 2000;53:88-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 59] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 23. | Chen CJ, Yu MW, Liaw YF. Epidemiological characteristics and risk factors of hepatocellular carcinoma. J Gastroenterol Hepatol. 1997;12:S294-S308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 338] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 24. | Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109:1266-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 114] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 25. | Ladero JM, Agúndez JA, Rodríguez-Lescure A, Diaz-Rubio M, Benítez J. RsaI polymorphism at the cytochrome P4502E1 locus and risk of hepatocellular carcinoma. Gut. 1996;39:330-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Rumack BH. Acetaminophen hepatotoxicity: the first 35 years. J Toxicol Clin Toxicol. 2002;40:3-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 27. | Johansson I, Eliasson E, Norsten C, Ingelman-Sundberg M. Hydroxylation of acetone by ethanol- and acetone-inducible cytochrome P-450 in liver microsomes and reconstituted membranes. FEBS Lett. 1986;196:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Nakajima T, Elovaara E, Park SS, Gelboin HV, Hietanen E, Vainio H. Immunochemical characterization of cytochrome P-450 isozymes responsible for benzene oxidation in the rat liver. Carcinogenesis. 1989;10:1713-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.0] [Reference Citation Analysis (0)] |