INTRODUCTION
Autologous and allogeneic tumor cells have been used as tumor vaccines in clinic for a long time[1,2]. The tumor cell-based vaccines possess the entire relevant tumor antigens recognized by the immune system and can be produced even without knowing the gene sequence of specific antigens. To identify the tumor specific antigen is time consuming, only few tumor specific antigens have been identified until now. But tumor cell-based immunization could not always elicit anti-tumor responses in sufficient magnitude to cause regression of established tumors due to scarcity of stimulatory surface molecules such as MHC-I, MHC-II or CD80[3].
Superantigens are a family of bacterial and viral proteins that bind to MHC class II molecules as unprocessed proteins and activate a large number of T cells bearing T cell receptor variable region beta chain (TCR Vβ)[4,5]. These T cells proliferate, and secrete cytokines (e.g. IFN-γ, TNF-α, IL-2, and IL-12), induce strong cytolytic activity, and mediate tumor regression[6-12]. SEA is an important member of superantigen family. Anchoring SEA onto MHC-II negative tumor cells through monoclonal antibody has been demonstrated to direct T cell-mediated cytotoxicity against these tumors[13,14]. Other studies suggeted that artificially anchoring a recombinant superantigen-transmembrane region chimera (SAg-TM) onto a tumor cell surface could substitute for the effect of MHC-II presentation[15].
In the present study we sought to construct an eukaryotic expression vector containing recombinant gene of SEA and transmembrane region sequence of CD80 molecule (CD80TM). Recombinant gene was transfected into HCC cells, chimera protein (SEA-CD80TM) was expressed on the membrane of living cells. SEA gene modified cells were irradiated and used as a superantigen enhanced tumor cell-based vaccine for HCC, which was capable of inducing antitumor immunity in vitro.
MATERIALS AND METHODS
Reagents
EX Taq DNA polymerase, T4 DNA ligation kit Ver.2.0, and restriction endonuclease were obtained from TakaRa Biotechnology (Dalian). DNA isolation and purification kit was purchased from Shanghai ShunHua Biotechnology. LipofectamineTM 2000 were from Invitrogen. IPTG, X-gal, DNA maker and FITC labeled sheep anti mouse IgG were from Sino-American Biotechnology. Dulbecco’s modified Eagle media, Trizol and G418 were from GibcoBRL. Access RT-PCR system was from Promega. Human INF-γ ELISPOT assay kit was from Diaclone, Recombinant human interleukin-2 (rhIL-2) and BD TriTEST™ antibody CD4 FITC/ CD8 PE/ CD3 PerCP were from BD Biosciences. Mouse anti-SEA monoclonal antibody was provided by the Department of Immunology, Fourth Military Medical University, P. R. China.
Plasmids
PBluescript II KS (+) plasmids containing cDNA of SEA and CD80 were constructed by Li et al[16]. Retroviral vector pLXSN was preserved in our laboratory.
Cell culture
Human HCC cell lines HepG2 and SMMC-7721 were cultured in DMEM containing 10% heat-inactivated fetal calf serum, 100 units/mL penicillin, 100 µg/mL streptomycin, 0.292 mg/mL glutamine.The cell lines were incubated in a humidified 5% CO2 incubator at 37 °C.
Polymerase chain reaction (PCR) and vector construction
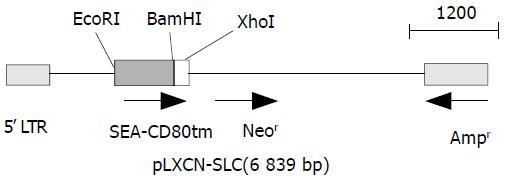
The primers used for cloning SEA (774 bp) and CD80TM (147 bp) were as follows. SEA, forward: GCGAATTCGCAT GAAAAAAACAGCATTTAC, reverse:CGGGATCCACTT GTATATAAATATATATCAAT. CD80TM, forward1: GGT GGCTCTGGCGGTGGCGGATCGGATAACCTGCTCCCATCC, forward2: CGGGATCCGGTGGAGGCGGTTCAGGCGG AGGTGGCTCTGGCGGT, reverse:CCGCTCGAGTTATAC AGGGCGTACACT. Linker (GGGGSGGGGSGGGGS) was introduced into the upstream of CD80TM using two forward primers. PCR was performed in a 50 µL reaction system consisting of 1 µM each primer, 200 µM each dNTP, 5 µL 10 × polymerase reaction buffer, and 1.25U EX Taq DNA polymerase with 1 µL SEA or CD80 gene vector. Samples were heated to 94 °C for 5 min followed by 30 cycles at 94 °C for 30 s, at 50 °C for 50 s, and at 68 °C for 1 min. The final extension was done at 72 °C for 7 min. Then 10 µL of each PCR product was electrophoresed on 1 % agarose gel containing 0.5 µg/mL EB, and PCR products were then purified from agarose gel according to the protocol of DNA purification kit. DNA fragments of SEA (S) and linker-CD80TM (LC) were subcloned into pBluescript II ks(+). The positive clones were selected from the transfected DH5α by the method described by Sambrook et al[17]. The constructed plasmids (designated as ks-S, ks-LC) were identified by restriction enzyme analysis. DNA sequences were verified in DNA sequencing core facility in Bioasia Biotechnology (Shanghai, China). Gene fragments were cut with EcoRI-BamHI, BamHI-XhoI, and inserted into the EcoRI-XhoI site of pLXSN to construct SAg gene expression vector pLXSN-SLC. The recombinant gene was verified by digestion with restriction endonuclease (Figure 1).
Figure 1 Gene map of expression vector.
In vitro transfection of pLXSN-SLC
pLXSN-SLC was transfected into HepG2 cells using LipofectamineTM 2000 reagent according to the manufacturer’s instructions. Briefly, 2 × 105 HepG2 cells were seeded into 12-well culture plates and cultured for 18 hours until 90% to 95% confluence of the cells, transfection was performed with 1 µg of DNA per dish. Forty eight h after transfection, cells were passaged at 1:4 dilution into a fresh medium. The next day, G418 400 µg/mL was added into culture medium for the screening of stably transfected cells. The cells were then grown in G418 culture medium for 4 weeks and cloned by limiting dilution. Mock cell line transfected with empty pLXSN vector was selected in G418, and used as control (HepG2-pLXSN).
Detection of SEA expression in transfected cells
Expression of SEA in screened monoclones was detected by RT-PCR. Total RNA was extracted from the tumor cells (HepG2 cells and SEA gene transfected or mock transfected HepG2 cells) using Trizol reagent. RNA (1.5 µg) was used to synthesize cDNA in 20 µL reaction mixtures following standard protocol of Access RT-PCR system. PCR was carried out with 5 µL of RT products in 50 µL PCR reaction buffer containing Tf1 polymerase, 1 mM MgCl2, annealing temperature was 50 °C. SEA cDNA was used as positive control in PCR. Indirect immunofluorescence was performed to locate the expressed SEA. Briefly, 2 × 105 SEA transfected HepG2 cells (HepG2-SEA) were plated into 12-well culture plates. At the bottom of each well a sterilized coverslip was placed in advance. HepG2 cells were plated as control. When the plated cells were 95%-100% confluence, the coverslips were washed twice in cold PBS (0.01M) and fixed with 4% paraformaldehyde for 30 min at room temperature. Cells were stained following the standard protocol of indirect immunofluorescence. First antibody was 1:100 diluted mouse anti-SEA monoclonal antibody in PBS, irrelevant sheep anti-mouse IgG was used as control. Second antibody was 1:500 diluted FITC labeled sheep anti-mouse IgG in 0.1‰ evan blue. Coverslips were then mounted directly onto a glass slide with a tiny drop of 50% glycerol in PBS (9.1 mM Na2HPO4/1.7 mM NaH2PO4/50 mM NaCl, pH 7.4). Fluorescent images were captured at 490 nm using a Nikon Eclipse E1000 microscope attached to a MicroMax camera (Princeton Instruments, Trenton, NJ).
Preparation of T cell lines
Human peripheral blood mononuclear cells (PBMC) were isolated from healthy volunteers by routine density centrifugation, and stimulated with HepG2-SEA to establish SEA-reactive T-cell line (SEA-T). The T cell line was stimulated repeatedly by exposure to irradiated (4 000 rad) HepG2-SEA and rhIL-2 (200 U/mL) in complete medium for 2 weeks, and then stored in frozen. These cells were thawed 1 week before use. T-cell lines stimulated by HepG2-pLXSN (pLXSN-T), HepG2 (HepG2-T) or just maintained with rhIL-2 (IL-2-T) were also prepared and used as control. Cell phenotypes of T cell lines were analyzed using BD TriTEST™ antibody by FACS.
Elispot assay
PVDF 96-well plates were incubated with 100 µL of 70% ethanol for 10 min at room temperature, and washed three times in 100 µL of PBS. Capture antibody was added and incubated at 4 °C overnight. After washed in 100 µL of PBS, each well was added 100 µL of 2% skimmed dry milk in PBS, and incubated for 2 hours at room temperature. Wells were emptied and tapped on absorbent paper, 5 × 104 effector cells (including SEA- T, pLXSN-T, HepG2-T, IL-2-T) and 1 × 104 target cells (irradiated HepG2) were seeded to each well and incubated for 20 h in RPMI -1640 medium without IL-2 and serum. Plates were then washed and incubated with detection antibody in PBS-1% BSA for 1 hour and 30 min at 37 °C, and with streptavidin-alkaline phosphatase for 1 h at 37 °C. After washed, substrate (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) was added and incubated for 5-20 min. After the final washing, dark-violet spots on plate membranes were counted under microscope.
Cytotoxicity of HepG2-SEA stimulated T cell line
Cytotoxicity was measured in a standard 4-hour 51Cr-release assay and expressed in formula: (%)specific lysis = 100 × (experimental - background cpm/maximal background cpm). Target cells (HepG2, SMMC-7721) were labeled for 2 hours with Na251CrO4 (51Cr, New England Nuclear, Boston, MA), 250 µCi/1 × 106 cells. The cells were then washed twice and 5 × 103 cells per well were seeded in triplicate in v-bottomed microtiter plates. Effector cells (SEA-T, HepG2-T and IL-2-T) were added at an effector-to-target (E:T) cell ratio of 40:1, 25:1, 10:1, 5:1 and 3:1, respectively. The plates were incubated at 37 °C and 5% CO2 for 4 h, supernatants were collected, and the released 51Cr was measured with a gamma counter (LKB-Wallac 1 282; Stockholm, Sweden). Spontaneous release was estimated by incubation of target cells in medium alone, and maximum release by resuspending the wells with 0.1% Tween 20. Spontaneous release was typically less than 30% of maximum release.
Statistical analysis
All experiments were performed in triplicate, data were presented as the mean ± SD and analyzed using Student’s t test.
RESULTS
Insertion of SEA and linker-CD80TM into MCS of pLXSN
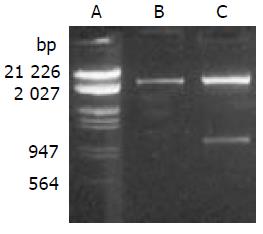
PCR products were subcloned into a pBluescript II ks (+) vector. DNA sequencing confirmed the correct sequence of both fragments. SEA gene was cloned into the EcoR I/Xho I site of pLXSN with linker-CD80TM. Recombinant gene SEA-linker-CD80TM was 1.0 kb with EcoR I and Xho I restriction site on each side, the size of pLXSN was 5.9 kb. Expressing vector pLXSN-SLC was digested by EcoR I/Xho I, 1.0 kb and 5.9 kb DNA fragments were appeared as expected (Figure 2).
Figure 2 Enzyme digestion analysis of expression vector.
A. Lambda DNA (EcoRI, HindIII); B. pLXSN (EcoRI, xhoI); C. pLXSN-SLC (EcoRI, xhoI).
SEA expressed on the membrane of HepG2
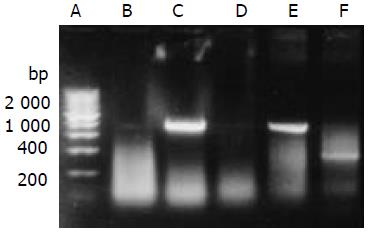
HepG2 was transfected with plasmid pLXSN-SLC and empty pLXSN vector, respectively. The expression of SEA was detected in HepG2 cells transfected with pLXSN-SLC only by RT-PCR (Figure 3). Indirect immunofluorescence test confirmed that most of the expressed SEA appeared to be located on the HepG2 cells membrane (Figure 4). SEA expression was detected in 5-10 percent of cells screened with G418. Irradiated HepG2-SEA cells were used as T cell stimulator in following experiments.
Figure 3 RT-PCR detected the transcription of SEA gene.
A. 200 bp DNA marker; B. HepG2; C. HepG2 transfected with pLXSN-SLC; D. HepG2 transfected with pLXSN; E. Positive control; F.β-actin (RNA of C).
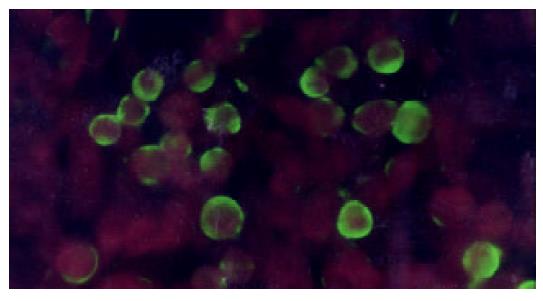
Figure 4 SEA expressed on the membrane of HepG2 (indirect immunofluorescence).
HepG2-SEA cells were stained accord-ing to the standard protocol of indirect immunofluorescence. First antibody is mouse anti-SEA IgG and second antibody is FITC labeled sheep anti-mouse IgG diluted in 0.1‰ evan blue. Positive signal is green while the background is red. SEA was located on the membrane of HepG2. Untransfected HepG2 cells did not express SEA (data not show).
Increased HepG2 reactive T cells with HepG2-SEA
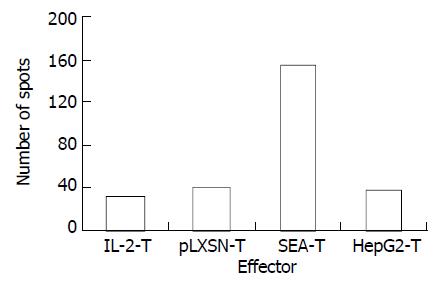
Interferon-γ (IFN-γ) ELISPOT assay was used to detect the specificity of HepG2 CD8+ T cells quantitatively. Cells tested included unstimulated PBMC (IL-2-T) and polyclonal T-lymphocyte lines stimulated by HepG2-SEA, HepG2-pLXSN or HepG2. After a short term of culture in vitro, the percentage of positive cells was more than 90% for CD3+, 80% to 90% for CD8+, and 5% to 10% for CD4+ (Figure 5). The number of HepG2 reactive CTL in the SEA-T cell line was much higher than that of HepG2-T (155.67 ± 7.22 vs 38.03 ± 8.18, t = 18.67, P < 0.0001), pLXSN-T (39.50 ± 6.26, t = 21.06, P < 0.0001) or IL-2-T (32.00 ± 4.26, t = 25.56, P < 0.0001) (Figure 6). Both the size and density of spots in SEA-T group were greater than those of controls (data not shown). The difference of spot number between HepG2-T and IL-2-T cell lines was not significant (t = -1.13, P = 0.321).
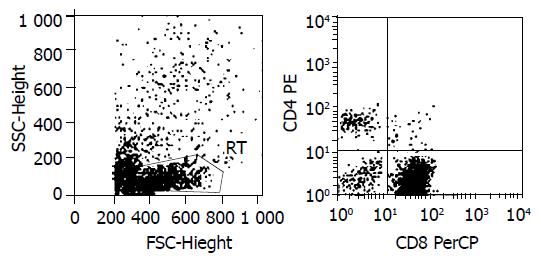
Figure 5 Lymphocyte phenotype analysis.
1 × 105 SEA-T cells were stained with BD TriTEST™ antibody CD4 FITC/CD8 PE/ CD3 PerCP, and assayed by FACS. CD3+90.75%, CD8+84.46%, CD4+6.29%.
Figure 6 HepG2-SEA increased the number of hepG2 specific CTL in T lymphocyte line.
Each bar represents average of spots in triplicate wells.
Enhanced cytotoxicity elicited by membrane expressed SEA
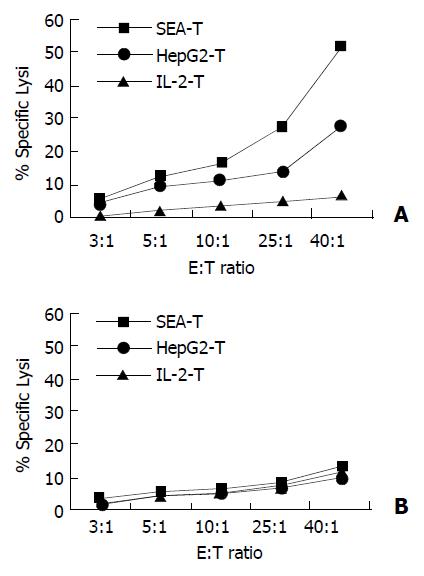
Cytotoxicity of the SEA-T effector cell line against HepG2, and SMMC-7721 target cells was measured (Figure 7). HepG2-T and IL-2-T cells were used as control. In contrast, SEA-T was highly efficient in directing specific lysis of HepG2 cells when compared with HepG2-T (51.55 ± 5 051% vs 33.16 ± 4.54, t = 4.47, P = 0.011). The two T cell lines had effective cytotoxicity to SMMC-7721 cells.
Figure 7 Membrane expressed SEA enhanced specific lysis on autologous tumor cells.
Cytotoxicity was measured in a stan-dard 4-hour 51Cr-release assay. Target cells (HepG2, SMMC-7721) were seeded in triplicate in v-bottomed microtiter plates at a concentration of 5 × 103 cells per well. Effector cells were added at an effector-to-target (E:T) cell ratio of 40:1, 25:1, 10:1, 5:1 and 3:1 respectively. A: SEA-T was highly efficient in di-recting specific lysis of HepG2 cells (squares), HepG2-T has moderate cytotoxicity on HepG2 (cross) cells (P < 0.001). B: None of T cell line has killing effect on SMMC-7721 cells.
DISCUSSION
T cells have been found to possess various potent antitumor effects through releasing cytotoxic effector molecules such as perforin or growth-suppressive cytokines IFN-γ and TNF-α[18-21]. However, the frequency of tumor-specific T cells could is generally too low and insufficient to interfere with progressive tumor growth. The goal of this experiment was to examine the possibility of developing a new experimental vaccine from irradiated HepG2 cells that were transfected to express superantigen-SEA on membrane (designed as HepG2-SEA) to augment cellular and humoral anti-tumor immune responses. Superantigen is the most effective T cell activator. Upon stimulation by superantigens, naïve T cells could respond and then become quickly anergized and/or deleted, while mature T cells could not become anergized[22-24]. Characteristis of superantigens can thus be exploited to enhance specific antigen responses. Because superantigens can cause anergy and/or deletion of potentially competing naïve T cells bearing the same Vβ element (s), thus there is less “competition” for cytokines and the desired specific immune response can be amplified.
HCC cells do not induce an effective immunological rejection process because of their weak antigenicity, the abnormal expression of MHC molecules as well as the deletion or deficiency of costimulatory molecules. In the present study, the expressed superantigen-SEA on the membrane of HepG2 cells was confirmed by RT-PCR and indirect immunofluorescence assays. When HepG2-SEA cells were used as a vaccine, two stimulating signals would present to T cells simultaneously, one was SEA, the other was tumor-associated antigens expressed on the surface of HepG2 cells, which facilitated the occurrence of specific anti-tumor immune reaction.
IFN-γ ELISPOT (enzyme-linked immunospot) assay was used for the quantitative detection of HepG2-specific CD8+ T cells. This technique could detect T cells that secrete a given cytokine (e.g., IFN-γ) in response to an antigenic stimulation[25]. The size and density of spots represented the quantity of IFN-γ secreted by single CTL[18]. The results showed that the number of HepG2 cell specific CTLs in repeatedly stimulated T cell line (SEA-T) with new vaccines was significantly increased than that of HepG2 cell-based vaccines (P < 0.001). Cytotoxic assay demonstrated that SEA-T was highly efficient in directing specific lysis of HepG2 cells other than SMMC-7721 cells. When compared with HepG2-T, the difference induced by new vaccines had statistical significance (P = 0.011). So we conclude that tumor cell membrane expressed SEA can stimulate the proliferation of tumor specific CTL and enhance the specific lysis on autologous tumor cells.
This superantigen modified tumor cell-based vaccine has an advantage over ordinary tumor cell-based vaccines in three aspects. First, this vaccine could make up for the disadvantage of low-level expression of class II MHC molecule on the surface of most tumor cells[26]. It has been found that T cell activation by SEA superantigen could occurr in the absence or presence of MHC class II[14,27-29], and artificially anchoring a superantigen onto a cell surface through transmembrane protein could substitute for MHC-II presentation[15]. Second, although most immunotherapeutic strategies have focused on activating tumor-specific CD8+ T cells, optimal antitumor activity could be achieved if both CD4+ and CD8+ tumor specific T cells were induced[30,31]. Likewise, if tumor immunity plays a role in limiting the recurrence of primary tumor or the future onset of metastatic diseases, then CD4+ and CD8+ immunological memory should be optimized. Both CD4+ and CD8+ T cells can be activated by bacterial superantigens. Third, superantigens could preferentially direct cytotoxicity against MHC-II-positive cells[32-34], in vivo administration of intact superantigens in sufficiently therapeutic amount could produce unwanted cytotoxicity to normal cells. Anchoring of superantigen and other TM fusion proteins on the surface of tumor cells would offer a novel strategy for the use of superantigens as immunostimulatory molecules. This method can confine the effect of SEA within the tumor, thus decreasing the side effects of treatment.
As presented above, irradiated HepG2-SEA cells could be used as an effective tumor cell-based vaccine in vitro. The superantigen enhanced vaccine is superior to tumor cell alone on the prime of tumor cell specific T cells, which provides a new strategy for the addition of superantigen as immunostimulatory molecules in tumor therapy. In vivo studies are currently under way to evaluate the effectiveness of these superantigen based vaccines for cancer therapy.