Published online Oct 1, 1995. doi: 10.3748/wjg.v1.i1.21
Revised: June 20, 1995
Accepted: August 12, 1995
Published online: October 1, 1995
AIM: To evaluate the role and analyze the loss of heterozygosity (LOH) of adenomatous polyposis coli (APC), mutation in colorectal cancer (MCC) and deleted in colorectal cancer (DCC) genes in the development and progression of colorectal cancers.
METHODS: LOH at APC, MCC and DCC genes was examined in 41 surgically resected specimens of colorectal carcinomas by polymerase chain reaction and restriction fragment length polymorphism analysis technique.
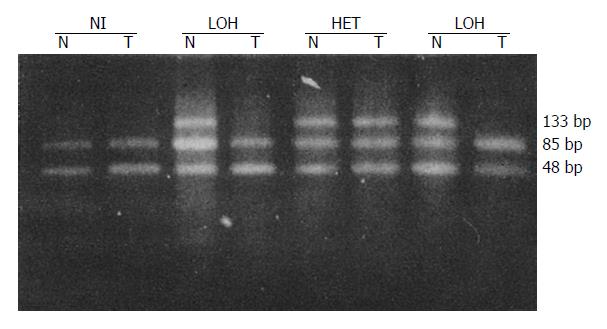
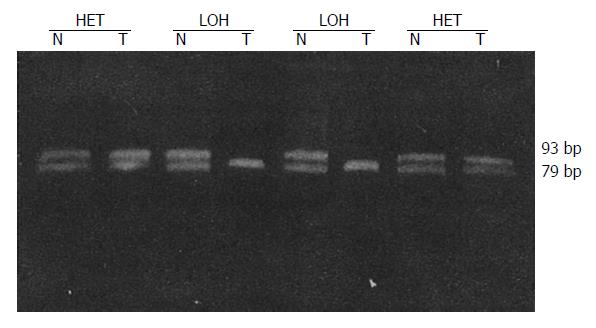
RESULTS: LOH of APC and MCC were observed in 7 of 25 (28.0%) and 8 of 22 (36.4%) of informative cases, respectively. When considered as one locus, the LOH frequency for APC/MCC was 14 of 36 (38.9%). LOH at DCC gene locus was detected in 21 of 38 (55.3%) of informative cases. No correlation was found between the LOH at APC or MCC gene and tumor histological types, size, invasion, lymph node metastasis and Dukes’ stages (P > 0.05). However, LOH rates at DCC locus in the group with lymph-node metastasis (80.0%) and in Dukes’ stages III and IV (71.4%) were significantly higher than those without lymph node metastasis (39.1%) and in Dukes’ stages I and II (35.3%) (P < 0.05).
CONCLUSION: LOH at APC and/or MCC may occur more frequently in the early stages and plays a role in the initiation of colorectal cancer while LOH at DCC is frequent at late event and associated with the progression and metastasis of colorectal cancer.
- Citation: Fang DC, Luo YH, Lu R, Liu WW, Liu FX, Liang ZY. Loss of heterozygosity at adenomatous polyposis coli, mutation in colorectal cancer and deleted in colorectal cancer genetic loci in colorectal cancers. World J Gastroenterol 1995; 1(1): 21-24
- URL: https://www.wjgnet.com/1007-9327/full/v1/i1/21.htm
- DOI: https://dx.doi.org/10.3748/wjg.v1.i1.21
Inactivation of tumor suppressor genes has been shown to play an important role in the development of a variety of human cancers[1,2]. The mechanisms of inactivation include allelic deletion, chromosome rearrangement, point mutation, and binding of suppressor gene products with viral or cellular inactivating proteins[1-3]. To date, several tumor suppressor genes have been discovered which include, but are not limited, the retinoblastoma susceptibility, p53, Wilm’s tumor, neurofibromatosis type I, adenomatous polyposis coli (APC), mutation in colorectal cancer (MCC), and deleted in colorectal cancer (DCC) genes. In this study, the loss of heterozygosity (LOH) at APC, MCC, and DCC genetic loci was further examined and analyzed.
Matching normal and tumor tissues were obtained at the time of surgery from 41 patients with colorectal carcinoma (11 with colonic carcinoma and 30 with rectal carcinoma). Each specimen was frozen immediately and stored at 80 oC until the analysis. A 5-μm section was cut from each tissue and stained with hematoxylin/cosin to ascertain whether the cancer cells in the tissues were predominant or not. Samples containing no cancer cells were considered normal, and those containing > 70% cancer cells were characterized as cancer-cell rich. Genomic DNA extraction was performed as previously described[4].
Polymerase chain reaction (PCR) was carried out as described previously[5]: 50 ng to 500 ng of genomic DNA were incubated at 95 oC for 5 min in 20 μL buffer containing 10 mmol/L Tris HCl, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 25 pmol/L of each primer, 200 μmol/L concentration of each deoxynucleotide triphosphate, and 2 units of Taq DNA polymerase. Multiple primer sets were used for each loci (Table 1). The priming regions were located within specific tumor suppressor genes of a polymorphic sequence[6]. Either a restriction fragment length polymorphism analysis technique (RFLP)[7] or a variable number of tandem repeats type polymorphism[8] was utilized. PCR was performed under the conditions described by Boynton et al[4] with a thermal cycler (Perkin Elmer Cetus, TOWN, COUNTRY). Annealing temperature, extension time and the number of amplification cycles were optimized for each primer set. PCR products were either digested with appropriate restriction enzymes (for RFLPs) or left intact (for a variable number of tandem repeats) and electrophoresed on either a 3% agarose gels or 8% polyacrylamide gel which were stained with ethidium bromide and photographed under UV light.
| Primer set1 | Priming region | Amplification size (base pairs) | Polymorphism type | Primer sequence |
| 1 | APC exon 11 | 133 | Rsal RFLP | 5'-GGACTACAGGCCATTGCAGAA-3' |
| 5'-GGCTACATCTCCAAAAGTCAA-3' | ||||
| 2 | MCC exon 10 | 79 or 93 | Insertion | 5'-TACGAATCCAATGCCACA-3' |
| 5'-CTGAAGTAGCTCCAAACA-3' | ||||
| 3 | DCC | 396 | MspI RFLP | 5'-TTGCACCATGCTGAAGATTGT-3' |
| 5'-ACCCTCCCCCTGATGACTTA-3' | ||||
| 4 | DCC | 240 | MspI RFLP | 5'-CGACTCGATCCTACAAAATC-3' |
| 5'-TCTACCCAGGTCTCAGAG-3' | ||||
| 5 | DCC | 200 | VNTR | 5'-GATGACATTTTCCCTCTAG-3' |
| 5'-GTGGTTATTGCCTTGAAAAG-3' |
LOH was defined as a visible change in allele: allele ratio in DNA relative to the ratio in corresponding normal DNA.
LOH results obtained for each locus are in Table 2. Tissues from 41 patients were studied for LOH. When multiple polymorphic loci within each gene were used, a constitutional heterozygosity (informativity) was found at APC in 25 (60.9%), at MCC in 22 (53.7%) and DCC in 38 cases (92.7%). LOH of APC and MCC were observed in 7 of 25 (28.0%) and 8 of 22 (36.4%) of informative cases, respectively (Figures 1 and 2). When considered as a single locus, the LOH frequency for APC/MCC (number positive for LOH of one or both genes/number informative for one or both genes) was 38.9% (14/36). LOH at DCC genetic locus was detected in 55.3% (21/38) informative cases (Figure 3, Figure 4, Figure 5). LOH of at least one of these three genes was detected in 68.3% (28/41) of tumor informative at all loci.
| Number | APC exon 11 | MCC exon 10 | DCC |
| 1 | HET | NI | NI |
| 2, 6, 12, 23 | HET | NI | HET |
| 3, 5, 7, 30, 32 | HET | NI | LON |
| 4 | LOH | HET | NI |
| 8, 19, 24, 25, 31 | NI | HET | LOH |
| 9, 11, 21 | HET | LOH | LOH |
| 10, 28, 37 | HET | HET | HET |
| 13 | LOH | LOH | HET |
| 14, 33, 40 | LOH | NI | HET |
| 15, 22, 34, 38 | NI | NI | LOH |
| 16, 20, 39 | NI | HET | HET |
| 17 | NI | LOH | NI |
| 18, 29 | NI | LOH | HET |
| 26 | HET | LOH | HET |
| 27 | HET | HET | LOH |
| 35 | LOH | NI | LOH |
| 36 | LOH | HET | LOH |
| 41 | NI | NI | HET |
Correlations between LOH at various loci and clinical pathological data of colorectal cancer are illustrated in Table 3. No significant correlation was found between the LOH at APC or MCC and tumor histological type, size, serosal invasion, lymph node metastases and Dukes’ stages (P > 0.05). However, the LOH rates at DCC locus in the groups with lymph node metastases and in the Dukes’ stages III and IV were significantly higher than in groups without lymph node metastases and in Dukes’ stages I and II (P < 0005).
| Group | LOH/ Informative (%) | ||
| APC | MCC | DCC | |
| Grade (differentiation) | |||
| Well/Moderate | 4/14 (28.6) | 4/12 (33.3) | 8/20 (40.0) |
| Low differentiated | 2/8 (25.0) | 3/6 (50.0) | 10/14 (71.4) |
| Mucoid | 1/3 (33.3) | 1/4 (25.0) | 3/4 (75.0) |
| Size | |||
| ≤ 3 cm | 3/8 (37.5) | 4/12 (33.3) | 7/15 (46.6) |
| > 3 cm | 4/17 (23.5) | 4/10 (40.0) | 14/23 (60.9) |
| Serosal invasion | |||
| Negative | 5/16 (37.5) | 4/14 (28.5) | 10/23 (43.4) |
| Positive | 2/9 (22.2) | 4/9 (50.0) | 11/15 (73.3) |
| Lymph-node metastasis | |||
| Negative | 4/16 (25.0) | 5/14 (35.7) | 9/23 (39.1) |
| Positive | 3/9 (33.3) | 3/8 (37.5) | 12/15 (80.0)a |
| Dukes' stages | |||
| Stages I and II | 4/14 (28.5) | 4/10 (40.0) | 6/17 (35.3) |
| Stages III and IV | 3/11 (27.3) | 4/12 (33.3) | 15/21 (71.4) |
Growing evidence suggests that the accumulation of multiple genetic events is responsible for the pathogenesis and/or progression of tumors. Multiple chromosomal deletions have been identified in colorectal cancer[9]. Recent studies on APC, MCC, and DCC gene aberrations have suggested that these genes may be involved in the carcinogenesis of human colorectal carcinoma[9-11]. LOH on chromosome 5q, where the APC and MCC genes are located, has been detected in 40.0% of sporadic colorectal carcinomas[9] and 33.0% of cancerous ulcerative colitis[11]. LOH on chromosome 18q, where the DCC gene is located, has been detected in 45.5% of sporadic colorectal carcinoma[9]. In the present study, LOH at APC and /or MCC was detected in 38.9% of colorectal carcinomas (APC, 28.0%; MCC, 36.4%), and at DCC in 55.3% of cases. These data suggest that deviations in APC, MCC and DCC genes may play a crucial role in the development and progression of colorectal carcinoma.
Genetic alterations such as ras mutation, 5q, 18q, and 17p deletions are believed to contribute to multistage carcinogenesis through colorectal adenoma to carcinoma[12]. LOH on 5q was observed most frequently in the intramucosal carcinoma[10,13]. With respect to the LOH on 18q, the frequency was very low in moderate and severe adenomas and intramucosal carcinomas, but it was high in invasive carcinomas[14]. In the present study, we did not find any correlation between LOH at APC and/or MCC and tumor histological type, size, serosal invasion, lymph node metastasis or the Dukes’ stages. However, the LOH rates at DCC locus in groups with lymph node metastasis and the Dukes’ stages III and IV were significantly higher than in groups without lymph node metastasis and the Dukes’ stages I and II. These data suggest that LOH at APC and/or MCC may occur more frequently in the early stages and play a role in the initiation of colorectal cancer. LOH at DCC is frequently a late event and is associated with the progression and metastasis of colorectal carcinoma.
Unexpectedly, there was no significant correlation between LOH of APC and MCC, even though these are closely linked loci on chromosome 5q. This finding suggests that LOH of APC occurs independently of LOH involving MCC. Similar discrepancies between these two genes have been previously reported in lung cancer[15] and esophageal cancer[4], as well as in colorectal cancer[16].
Some tumors did not lose heterozygosity at any of the tumor suppressor gene loci examined. One explanation is that these genes may be altered by another mechanism, such as point mutation, gene rearrangement, or microdeletion. Point mutations in APC, MCC, and DCC have been found in tumors without LOH[16]. Another explanation is that LOH at chromosomal regions, such as chromosomal 17q, is also important in pathogenesis. Furthermore, current assays may have limited sensitivity due to normal cell contamination of the specimens and/or lack of informativity at the RFLPs tested. And lastly, a subset of colorectal tumors may arise through the inactivation of other, as yet unknown, tumor suppressor genes and/or in combination with other genetic and epigenetic events. Further studies are required to explore these possibilities.
Original title: China National Journal of New Gastroenterology (1995-1997) renamed World Journal of Gastroenterology (1998-).
S- Editor: Filipodia L- Editor: Jennifer E- Editor: Zhang FF
| 1. | Weinberg RA. Tumor suppressor genes. Science. 1991;254:1138-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1050] [Cited by in RCA: 988] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 2. | Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2565] [Cited by in RCA: 2682] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 3. | Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1398] [Cited by in RCA: 1445] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 4. | Boynton RF, Blount PL, Yin J, Brown VL, Huang Y, Tong Y, McDaniel T, Newkirk C, Resau JH, Raskind WH. Loss of heterozygosity involving the APC and MCC genetic loci occurs in the majority of human esophageal cancers. Proc Natl Acad Sci USA. 1992;89:3385-3388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 144] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Gao X, Honn KV, Grignon D, Sakr W, Chen YQ. Frequent loss of expression and loss of heterozygosity of the putative tumor suppressor gene DCC in prostatic carcinomas. Cancer Res. 1993;53:2723-2727. [PubMed] |
| 6. | Tamura G, Maesawa C, Suzuki Y, Ogasawara S, Terashima M, Saito K, Satodate R. Primary gastric carcinoma cells frequently lose heterozygosity at the APC and MCC genetic loci. Jpn J Cancer Res. 1993;84:1015-1018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314-331. [PubMed] |
| 8. | Nakamura Y, Leppert M, O’Connell P, Wolff R, Holm T, Culver M, Martin C, Fujimoto E, Hoff M, Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987;235:1616-1622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1090] [Cited by in RCA: 1003] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 9. | Iino H, Fukayama M, Maeda Y, Koike M, Mori T, Takahashi T, Kikuchi-Yanoshita R, Miyaki M, Mizuno S, Watanabe S. Molecular genetics for clinical management of colorectal carcinoma. 17p, 18q, and 22q loss of heterozygosity and decreased DCC expression are correlated with the metastatic potential. Cancer. 1994;73:1324-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 10. | Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P, Markham A. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 512] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 11. | Greenwald BD, Harpaz N, Yin J, Huang Y, Tong Y, Brown VL, McDaniel T, Newkirk C, Resau JH, Meltzer SJ. Loss of heterozygosity affecting the p53, Rb, and mcc/apc tumor suppressor gene loci in dysplastic and cancerous ulcerative colitis. Cancer Res. 1992;52:741-745. [PubMed] |
| 12. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4616] [Cited by in RCA: 4464] [Article Influence: 120.6] [Reference Citation Analysis (0)] |
| 13. | Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1920] [Cited by in RCA: 1863] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 14. | Miyaki M, Seki M, Okamoto M, Yamanaka A, Maeda Y, Tanaka K, Kikuchi R, Iwama T, Ikeuchi T, Tonomura A. Genetic changes and histopathological types in colorectal tumors from patients with familial adenomatous polyposis. Cancer Res. 1990;50:7166-7173. [PubMed] |
| 15. | D’Amico D, Carbone DP, Johnson BE, Meltzer SJ, Minna JD. Polymorphic sites within the MCC and APC loci reveal very frequent loss of heterozygosity in human small cell lung cancer. Cancer Res. 1992;52:1996-1999. [PubMed] |
| 16. | Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1276] [Cited by in RCA: 1247] [Article Influence: 36.7] [Reference Citation Analysis (0)] |