Copyright
©The Author(s) 2024.
World J Gastroenterol. Apr 14, 2024; 30(14): 2038-2058
Published online Apr 14, 2024. doi: 10.3748/wjg.v30.i14.2038
Published online Apr 14, 2024. doi: 10.3748/wjg.v30.i14.2038
Figure 1 Treatment with caerulein alone or in combination with lipopolysaccharide induced varying degrees of experimental acute pancreatitis.
Modeling methods for the control group, caerulein (CAE) group, and CAE combined with lipopolysaccharide (LPS; CAE + LPS) group, evaluation of pancreatic tissue changes, and relevant parameters. A: Schematic representation of the modeling methods for the CAE group and CAE + LPS group; B: Pancreatic weight ratios in the three groups; C: H&E staining of pancreatic tissue; D: Scores for edema, inflammation, and necrosis in pancreatic pathology; E-G: Quantitative real-time PCR expression values of pancreatic inflammatory factors. n = 6 per group, aP < 0.05, bP < 0.01, cP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
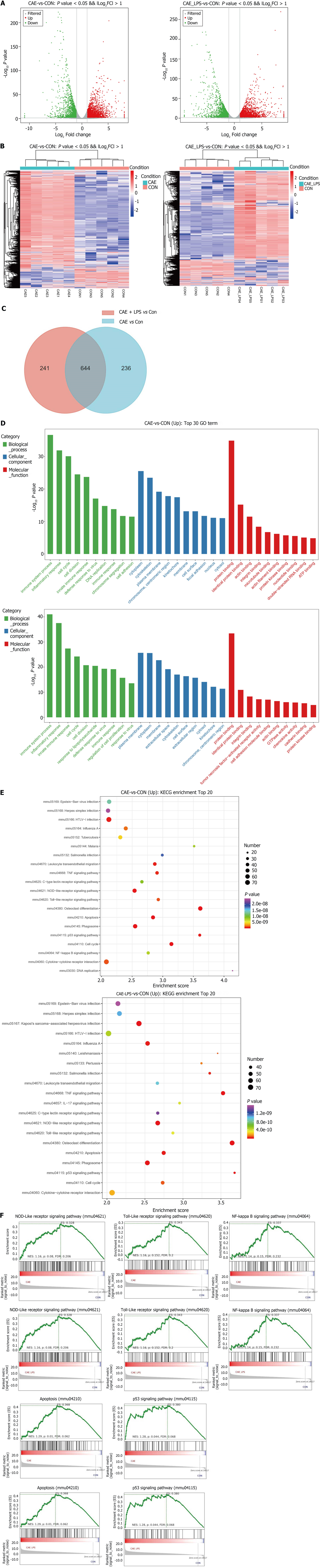
Figure 2 RNA-seq identified the molecular changes during the development of acute pancreatitis.
Comparative analysis of the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group with the control (Con) group. A: Volcano plot of significant differentially expressed (SDE) genes (P value < 0.05 and |log2FC| > 1), with red indicating significant upregulation and green indicating significant downregulation; B: Venn diagram, where blue represents CAE group vs Con group, and red represents CAE + LPS group vs Con group, with the gray area representing commonly differentially expressed genes in both groups compared to the Con group; C: Heatmap displaying the expression of SDE genes in each pancreatic sample, with red indicating upregulated genes and blue indicating downregulated genes; D: Gene Ontology (GO) functional annotation of the top 30 significantly different genes, with the GO classification chart showing the distribution of entries related to biological processes, cellular components, and molecular functions; E: Enrichment analysis results of the top 20 significantly different Kyoto Encyclopedia of Genes and Genomes pathways; F: Gene Set Enrichment Analysis for the NOD-like receptor, TLR, NF-κB, and P53 signaling pathways, as well as apoptosis. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
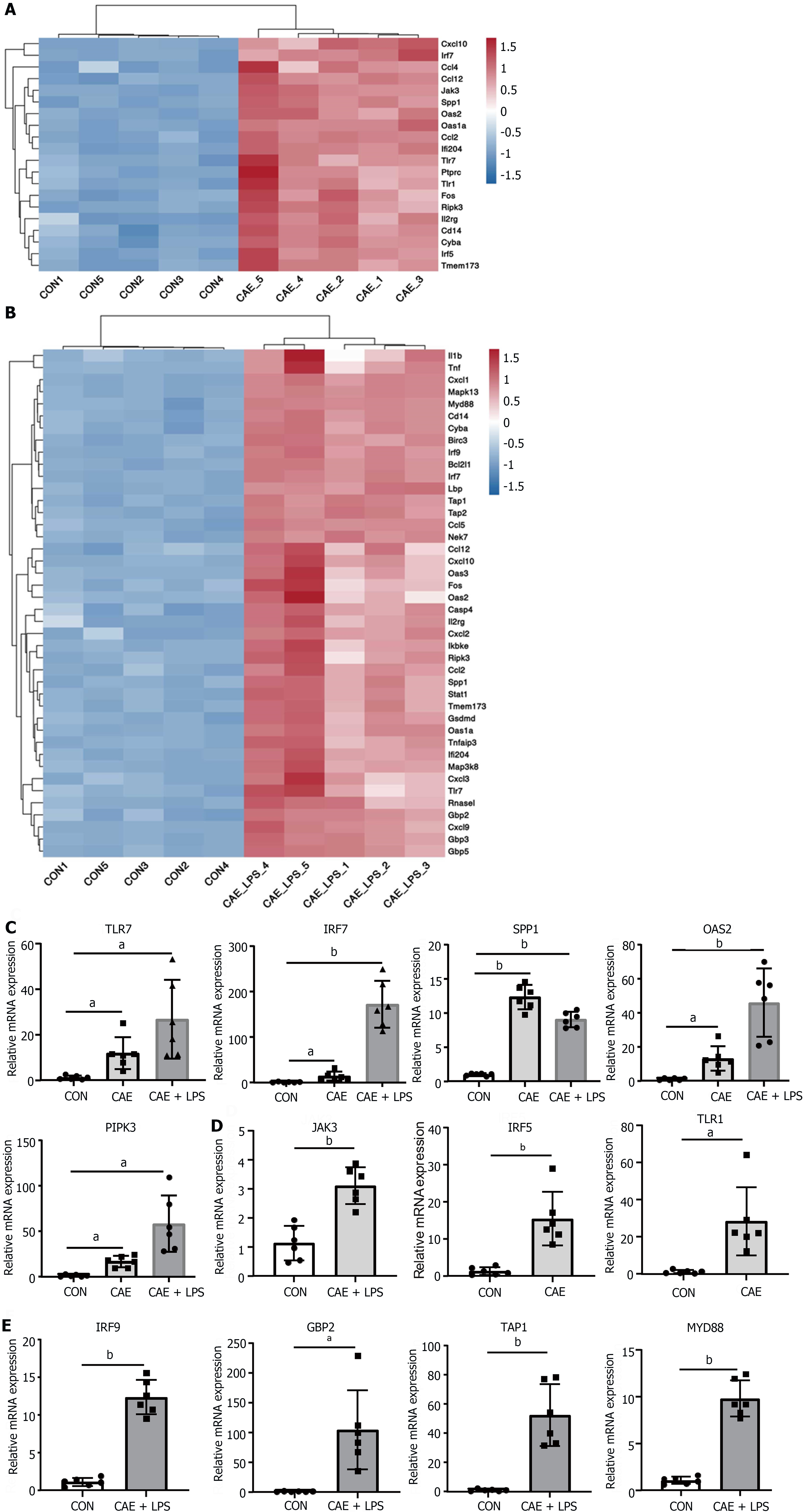
Figure 3 NOD-like receptor and TLR signaling pathway played a crucial role in the progression of acute pancreatitis.
Significant differentially expressed genes in the NOD-like receptor and TLR signaling pathways after comparing the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group to the control (Con) group. A: Heatmap of significant differentially expressed (SDE) genes in the CAE Group compared to the Con group; B: Heatmap of SDE genes in the CAE + LPS group compared to the Con group; C: Quantitative real-time PCR (qRT-PCR) expression values of commonly significantly different genes in both groups; D: qRT-PCR expression values of specifically significantly different genes in the CAE Group compared to the Con group; E: qRT-PCR expression values of specifically significantly different genes in the CAE + LPS group compared to the Con group. n = 6 per group, aP < 0.01, bP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
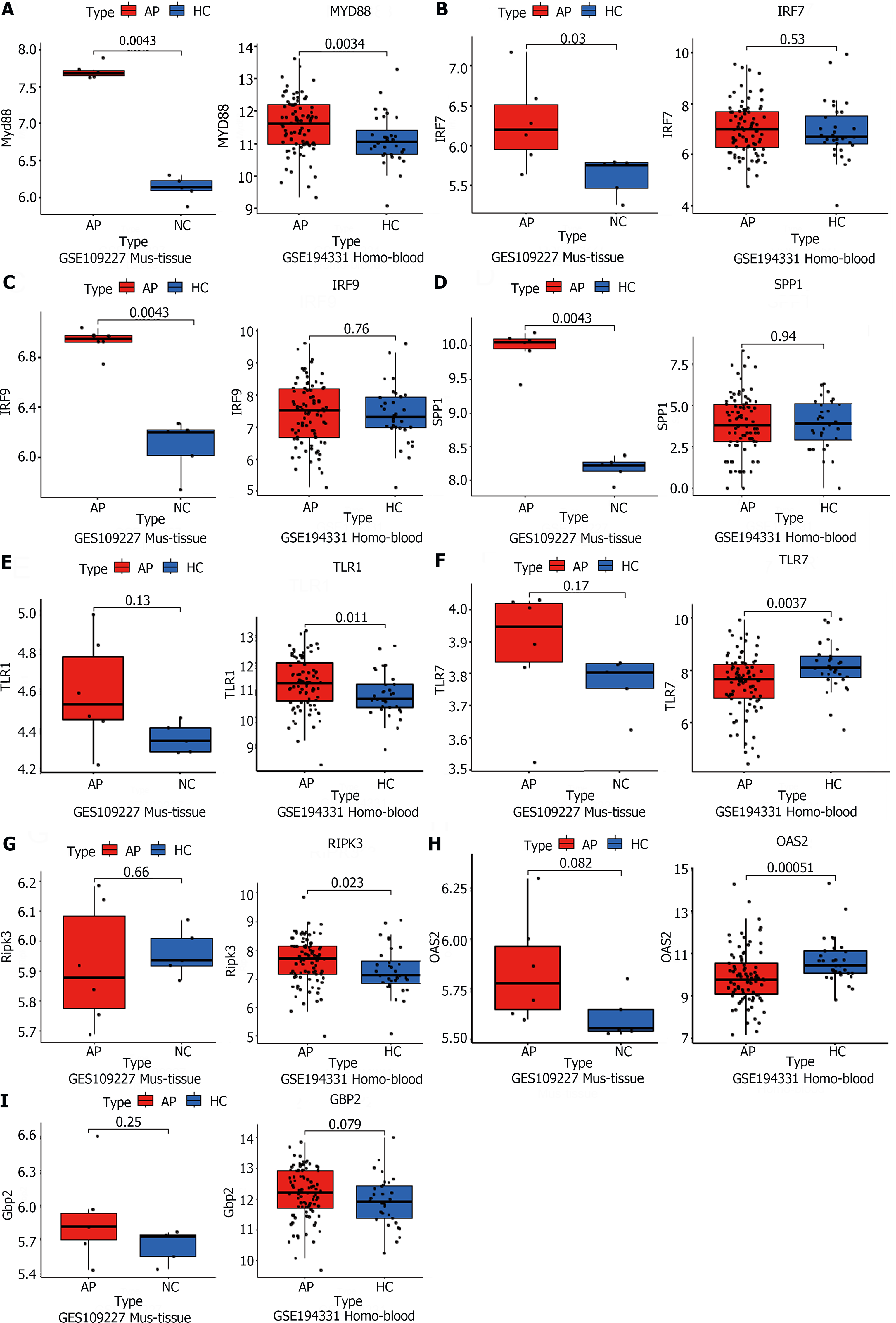
Figure 4 Public RNA-seq data reveals that the significant role of TLR and NOD-like signaling pathways in acute pancreatitis pathogenesis.
A-I: The acute pancreatitis (AP) animal RNA-seq dataset (GSE109227) and human AP patient blood RNA-seq dataset (GSE194331) were downloaded from the Gene Expression Omnibus database for the external validation of significant differentially expressed genes in the successfully validated NOD-like receptor and TLR signaling pathways in the animal model, using quantitative real-time PCR. n = 6 per group.
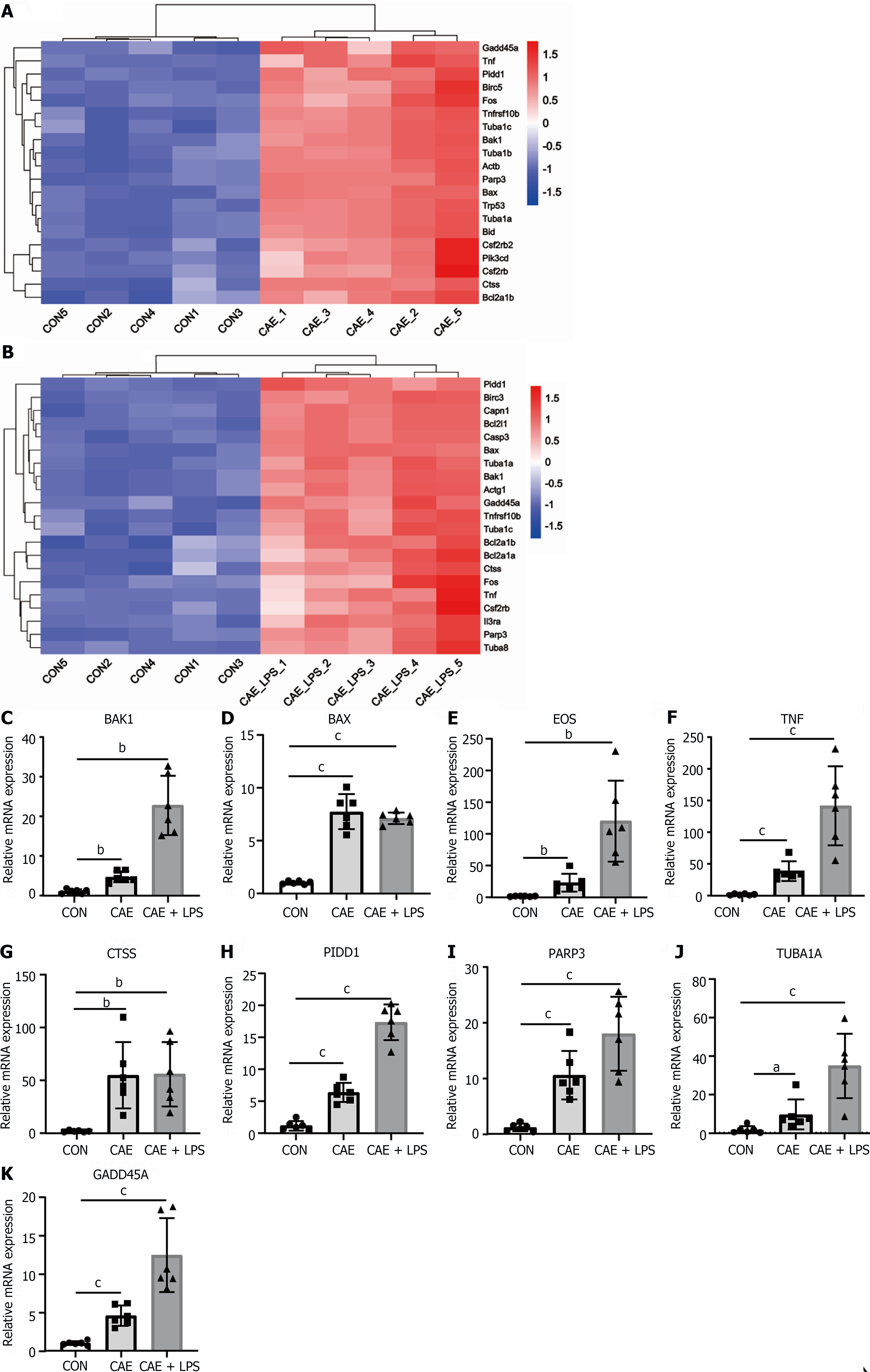
Figure 5 Apoptosis served as a crucial pathway mediating pancreatic necrosis in acute pancreatitis.
Significant differentially expressed (SDE) genes in the apoptotic signaling pathway after comparing the caerulein (CAE) group and CAE + lipopolysaccharide (LPS) group to the control (Con) group. A: Heatmap of SDE genes in the CAE group compared to the Con group; B: Heatmap of SDE genes in the CAE + LPS group compared to the Con group; C-K: Quantitative real-time PCR expression values of commonly significantly different genes in both groups, n = 6 per group, aP < 0.05, bP < 0.01, cP < 0.001. CAE: Caerulein; CON: Control; LPS: Lipopolysaccharide.
Figure 6 TUBA1A and GADD45A were genes closely associated with apoptosis in acute pancreatitis.
A-I: The acute pancreatitis (AP) animal RNA-seq dataset (GSE109227) and human AP patient blood RNA-seq dataset (GSE194331) were downloaded from the Gene Expression Omnibus database for the external validation of the genes successfully validated in the animal model and related to the apoptotic signaling pathway, using quantitative real-time PCR. AP: Acute pancreatitis; HC: Healthy control.
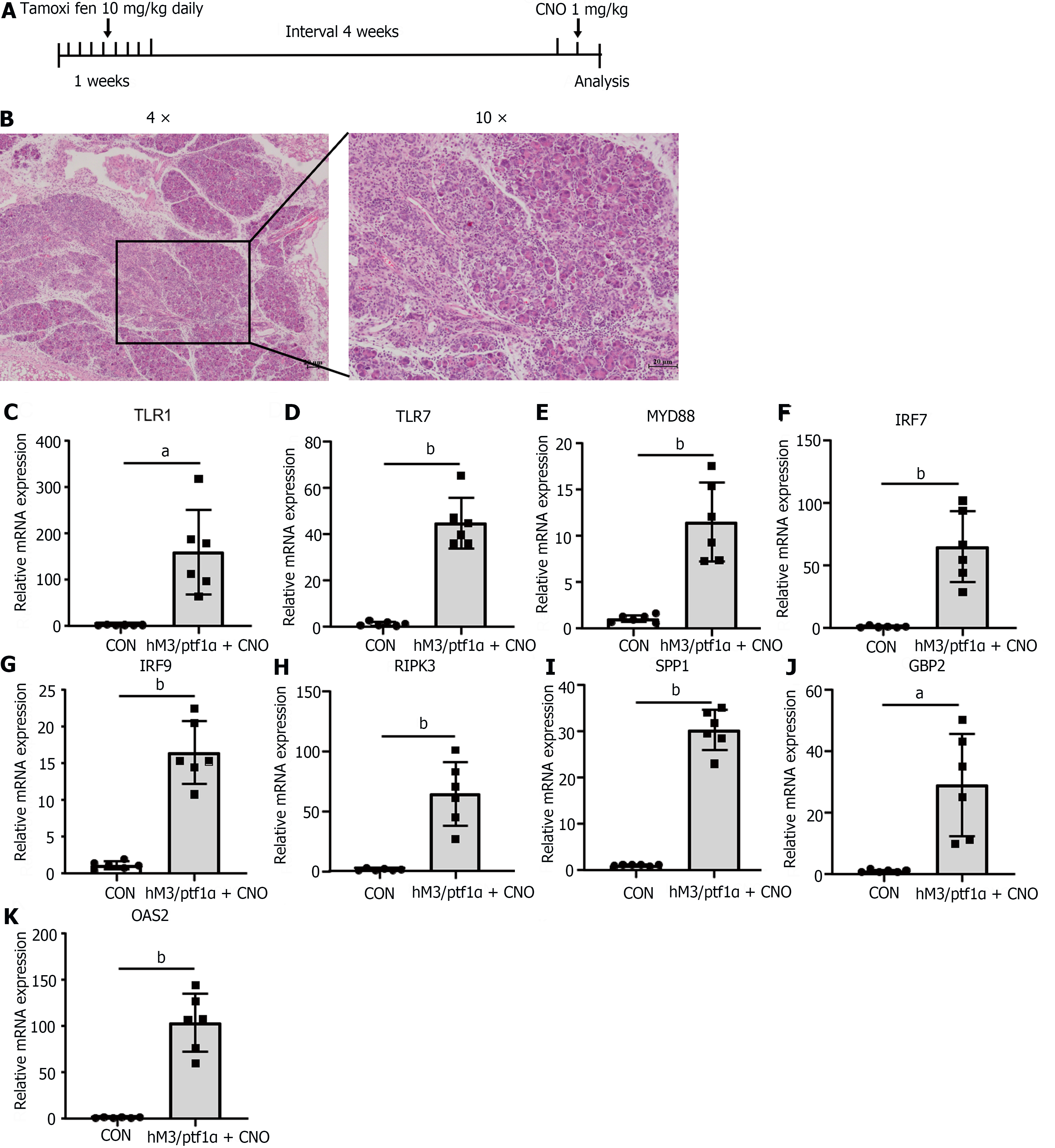
Figure 7 Inflammatory response in hM3/Ptf1α(cre) transgenic animal models consistent with the caerulein model.
hM3/Ptf1α(cre)model and its pathological images with inflammatory gene validation. A: Schematic representation of the hM3/Ptf1α(cre) model; B: H&E staining of pancreatic tissue schematic representation of the modeling methods; C-K: Validation of the significant differentially expressed genes in the transgenic animal hM3/Ptf1α(cre) model after inducing acute pancreatitis, which were found in the NOD-like receptor and TLR signaling pathways. n = 6 per group, aP < 0.01, bP < 0.001. CON: Control; CNO: Clozapine N-oxide.
Figure 8 The apoptosis in acute pancreatitis in the hM3/Ptf1α(cre) transgenic animal model was consistent with the caerulein models.
A-I: Validation of the significant differentially expressed genes in the NOD-like receptor and TLR signaling pathways in the transgenic animal hM3/Ptf1α(cre) model after inducing acute pancreatitis. n = 6 per group, aP < 0.01, bP < 0.001. CON: Control; CNO: Clozapine N-oxide.
- Citation: Zheng P, Li XY, Yang XY, Wang H, Ding L, He C, Wan JH, Ke HJ, Lu NH, Li NS, Zhu Y. Comparative transcriptomic analysis reveals the molecular changes of acute pancreatitis in experimental models. World J Gastroenterol 2024; 30(14): 2038-2058
- URL: https://www.wjgnet.com/1007-9327/full/v30/i14/2038.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i14.2038