Copyright
©The Author(s) 2022.
World J Gastroenterol. Dec 21, 2022; 28(47): 6689-6701
Published online Dec 21, 2022. doi: 10.3748/wjg.v28.i47.6689
Published online Dec 21, 2022. doi: 10.3748/wjg.v28.i47.6689
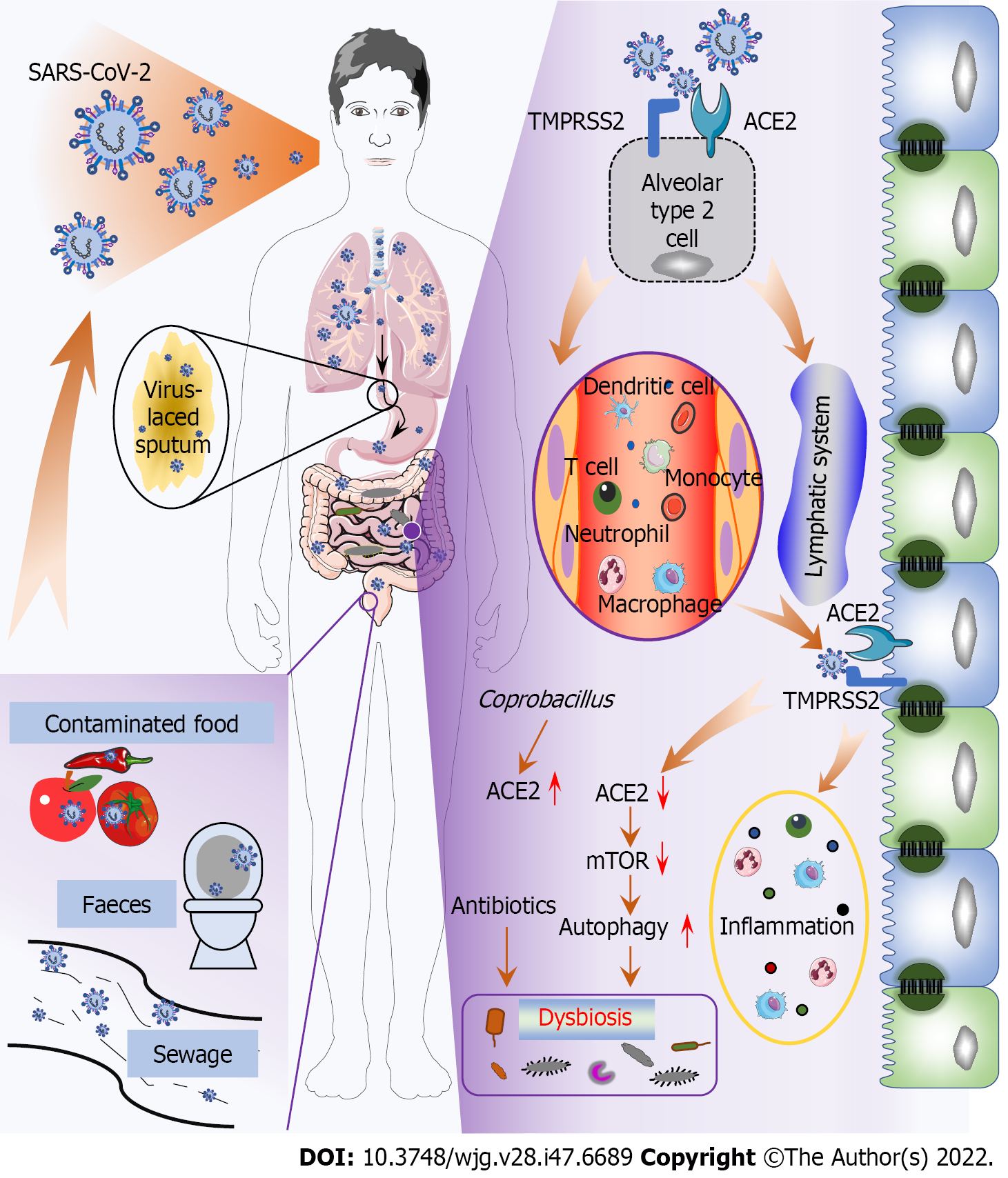
Figure 1 Potential pathways and mechanisms of gut infection and dysbiosis induced by severe acute respiratory syndrome coronavirus 2.
Fecal-oral transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated water and food, or by swallowing virus-laced sputum, results in gut infection and dysbiosis. SARS-CoV-2 infects alveolar type 2 cells and damages lung tissue before invading the gut through circulating immune cells and the lymphatic system and infiltrating gut epithelial cells via angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2, subsequently triggering gut inflammation and further promoting dysbiosis. Internalization of SARS-CoV-2 Leads to downregulation of ACE2, resulting in inhibition of mechanistic target of rapamycin and subsequent activation of gut autophagy, which modulates the gut microbiome. However, gut microbiota top associations with disease severity in coronavirus disease 2019 patients, such as Coprobacillus, have been shown to upregulate ACE2 expression in the gut. The improper use of antibiotics can also lead to dysbiosis. TMPRSS2: Transmembrane serine protease 2; ACE2: Angiotensin-converting enzyme 2; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; mTOR: Mechanistic target of rapamycin.
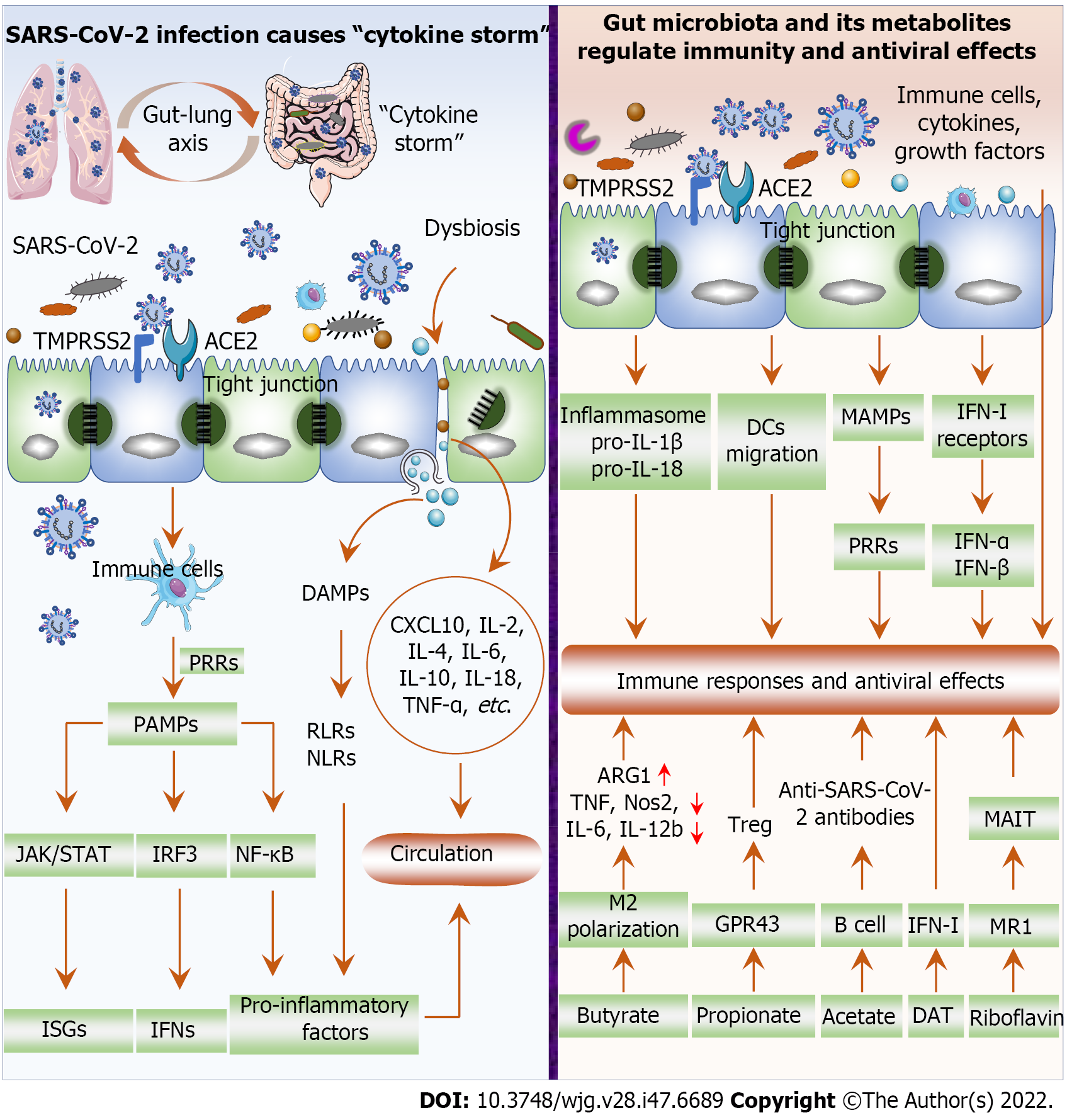
Figure 2 Severe acute respiratory syndrome coronavirus 2 infection causes “cytokine storm” and the impact of gut microbiota and its metabolites on inflammatory response and antiviral effects.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) invades cells through angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) and replicates frantically, inducing innate immune cells [e.g., macrophages, dendritic cells (DCs)] to recognize and bind pathogen-associated molecular patterns through pattern recognition receptors (PRRs). Subsequently, the expression of pro-inflammatory factors, interferons (IFNs) and IFN-stimulated genes is induced through the NF-κB, IRF3, and JAK/STAT signaling pathways, thereby promoting excessive inflammatory cytokine release. Killing or damaging cells by SARS-CoV-2 releases danger-associated molecular patterns that facilitate the expression of pro-inflammatory factors via activation of RIG-I-like receptors and NOD-like receptors. In addition, dysbiosis damages the gut barrier (tight junctions) and promotes the production of inflammatory factors such as CXCL10, IL-2, IL-4, IL-6, IL-10, IL-18, and TNF-α. Gut microbiota facilitates inflammasome activation, pro-IL-1β and pro-IL-18 expression, and DCs migration, thereby promoting protective immunity post viral infection. Microbial-associated molecular patterns are transmitted to the parenteral tissues to activate PRRs and affect innate immune responses. Gut microbiota regulates IFN-I receptors expression in respiratory epithelial cells and exerts antiviral effects through IFN-α and IFN-β. Immune cells, cytokines, and growth factors in the gut mucosa reach the respiratory tract to regulate immunity and exert antiviral effects. Butyrate promotes M2 macrophage polarization, upregulating arginase 1, and downregulating TNF, Nos2, IL-6, and IL-12b exerts anti-inflammatory activity. Propionate promotes Treg cell proliferation by activating G-protein-coupled receptor 43, thereby inhibiting autoinflammatory responses and protecting tissues from damage caused by pathological immune responses. Acetate promotes the production of SARS-CoV-2 antibodies by B cells, thereby inhibiting the development of COVID-19. The gut microbiota produces deaminotyrosine to enhance IFN-I signaling and protect the host from viral infection. Riboflavin, produced by the gut microbiome, activates mucosal-associated T cells via major histocompatibility complex-related protein-1. Mucosal-associated T cells participate in the immune response against SARS-CoV-2 through the gut-lung axis, thereby exerting antiviral efficacy. TMPRSS2: Transmembrane serine protease 2; ACE2: Angiotensin-converting enzyme 2; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; PRR: Pattern recognition receptors; IFN: Interferons; ISG: Interferons-stimulated gene; MAIT: Mucosal-associated T cells; MAMP: Microbial-associated molecular pattern; DAT: Deaminotyrosine; RLR: RIG-I-like receptors; NLR: NOD-like receptors; DAMP: Danger-associated molecular pattern; PAMP: Pathogen-associated molecular pattern; DC: Dendritic cell; MR1: Major histocompatibility complex-related protein-1; GPR43: G-protein-coupled receptor 43.
- Citation: Xiang H, Liu QP. Alterations of the gut microbiota in coronavirus disease 2019 and its therapeutic potential. World J Gastroenterol 2022; 28(47): 6689-6701
- URL: https://www.wjgnet.com/1007-9327/full/v28/i47/6689.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i47.6689